Exploring Gut Microbiome Composition and Circulating Microbial DNA Fragments in Patients with Stage II/III Colorectal Cancer: A Comprehensive Analysis
- PMID: 38792001
- PMCID: PMC11119035
- DOI: 10.3390/cancers16101923
Exploring Gut Microbiome Composition and Circulating Microbial DNA Fragments in Patients with Stage II/III Colorectal Cancer: A Comprehensive Analysis
Abstract
Background: Colorectal cancer (CRC) significantly contributes to cancer-related mortality, necessitating the exploration of prognostic factors beyond TNM staging. This study investigates the composition of the gut microbiome and microbial DNA fragments in stage II/III CRC.
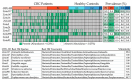
Methods: A cohort of 142 patients with stage II/III CRC and 91 healthy controls underwent comprehensive microbiome analysis. Fecal samples were collected for 16S rRNA sequencing, and blood samples were tested for the presence of microbial DNA fragments. De novo clustering analysis categorized individuals based on their microbial profiles. Alpha and beta diversity metrics were calculated, and taxonomic profiling was conducted.
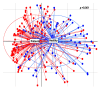
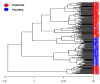
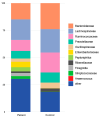
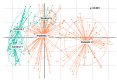
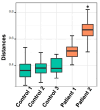
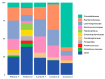
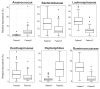
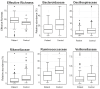
Results: Patients with CRC exhibited distinct microbial composition compared to controls. Beta diversity analysis confirmed CRC-specific microbial profiles. Taxonomic profiling revealed unique taxonomies in the patient cohort. De novo clustering separated individuals into distinct groups, with specific microbial DNA fragment detection associated with certain patient clusters.
Conclusions: The gut microbiota can differentiate patients with CRC from healthy individuals. Detecting microbial DNA fragments in the bloodstream may be linked to CRC prognosis. These findings suggest that the gut microbiome could serve as a prognostic factor in stage II/III CRC. Identifying specific microbial markers associated with CRC prognosis has potential clinical implications, including personalized treatment strategies and reduced healthcare costs. Further research is needed to validate these findings and uncover underlying mechanisms.
Keywords: colorectal cancer; gut microbiome; microbial DNA fragments; microbial dysbiosis; prognostic markers.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Fecal Microbiota and Gut Microbe-Derived Extracellular Vesicles in Colorectal Cancer.Front Oncol. 2021 Sep 14;11:650026. doi: 10.3389/fonc.2021.650026. eCollection 2021. Front Oncol. 2021. PMID: 34595105 Free PMC article.
-
Analysis of the gut microbiome using extracellular vesicles in the urine of patients with colorectal cancer.Korean J Intern Med. 2023 Jan;38(1):27-38. doi: 10.3904/kjim.2022.112. Epub 2022 Nov 10. Korean J Intern Med. 2023. PMID: 36353788 Free PMC article.
-
Gut microbiota dysbiosis signature is associated with the colorectal carcinogenesis sequence and improves the diagnosis of colorectal lesions.J Gastroenterol Hepatol. 2020 Dec;35(12):2109-2121. doi: 10.1111/jgh.15077. Epub 2020 Jun 22. J Gastroenterol Hepatol. 2020. PMID: 32337748
-
Gut Microbial and Associated Metabolite Markers for Colorectal Cancer Diagnosis.Microorganisms. 2023 Aug 8;11(8):2037. doi: 10.3390/microorganisms11082037. Microorganisms. 2023. PMID: 37630597 Free PMC article. Review.
-
Diet-mediated gut microbial community modulation and signature metabolites as potential biomarkers for early diagnosis, prognosis, prevention and stage-specific treatment of colorectal cancer.J Adv Res. 2023 Oct;52:45-57. doi: 10.1016/j.jare.2022.12.015. Epub 2022 Dec 31. J Adv Res. 2023. PMID: 36596411 Free PMC article. Review.
Cited by
-
Obesity-Associated Colorectal Cancer.Int J Mol Sci. 2024 Aug 14;25(16):8836. doi: 10.3390/ijms25168836. Int J Mol Sci. 2024. PMID: 39201522 Free PMC article. Review.
-
Circulating Bacterial DNA in Colorectal Cancer Patients: The Potential Role of Fusobacterium nucleatum.Int J Mol Sci. 2024 Aug 20;25(16):9025. doi: 10.3390/ijms25169025. Int J Mol Sci. 2024. PMID: 39201711 Free PMC article.
References
-
- Kohne C.H., Schoffski P., Wilke H., Kaufer C., Andreesen R., Ohl U., Klaasen U., Westerhausen M., Hiddemann W., Schott G., et al. Effective biomodulation by leucovorin of high-dose infusion fluorouracil given as a weekly 24-h infusion: Results of a randomized trial in patients with advanced colorectal cancer. J. Clin. Oncol. 1998;16:418–426. doi: 10.1200/JCO.1998.16.2.418. - DOI - PubMed
-
- Argente-Pla M., Perez-Lazaro A., Martinez-Millana A., Del Olmo-Garcia M.I., Espi-Reig J., Beneyto-Castello I., Lopez-Andujar R., Merino-Torres J.F. Simultaneous Pancreas Kidney Transplantation Improves Cardiovascular Autonomic Neuropathy with Improved Valsalva Ratio as the Most Precocious Test. J. Diabetes Res. 2020;2020:7574628. doi: 10.1155/2020/7574628. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

