Genotyping Hepatitis B virus by Next-Generation Sequencing: Detection of Mixed Infections and Analysis of Sequence Conservation
- PMID: 38791519
- PMCID: PMC11122360
- DOI: 10.3390/ijms25105481
Genotyping Hepatitis B virus by Next-Generation Sequencing: Detection of Mixed Infections and Analysis of Sequence Conservation
Abstract
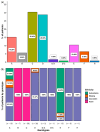
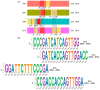
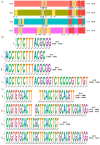
Our aim was to develop an accurate, highly sensitive method for HBV genotype determination and detection of genotype mixtures. We examined the preS and 5' end of the HBV X gene (5X) regions of the HBV genome using next-generation sequencing (NGS). The 1852 haplotypes obtained were subjected to genotyping via the Distance-Based discrimination method (DB Rule) using two sets of 95 reference sequences of genotypes A-H. In clinical samples from 125 patients, the main genotypes were A, D, F and H in Caucasian, B and C in Asian and A and E in Sub-Saharan patients. Genotype mixtures were identified in 28 (22.40%) cases, and potential intergenotypic recombination was observed in 29 (23.20%) cases. Furthermore, we evaluated sequence conservation among haplotypes classified into genotypes A, C, D, and E by computing the information content. The preS haplotypes exhibited limited shared conserved regions, whereas the 5X haplotypes revealed two groups of conserved regions across the genotypes assessed. In conclusion, we developed an NGS-based HBV genotyping method utilizing the DB Rule for genotype classification. We identified two regions conserved across different genotypes at 5X, offering promising targets for RNA interference-based antiviral therapies.
Keywords: Distance-Based discrimination method; RNA interference; conservation; genotypes; hepatitis B X gene; hepatitis B virus; information content; next-generation sequencing; preS; quasispecies.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Detection of hyper-conserved regions in hepatitis B virus X gene potentially useful for gene therapy.World J Gastroenterol. 2018 May 21;24(19):2095-2107. doi: 10.3748/wjg.v24.i19.2095. World J Gastroenterol. 2018. PMID: 29785078 Free PMC article.
-
Distribution of hepatitis B virus genotypes in the general population of Myanmar via nationwide study.BMC Infect Dis. 2020 Jul 29;20(1):552. doi: 10.1186/s12879-020-05269-z. BMC Infect Dis. 2020. PMID: 32727389 Free PMC article.
-
Analysis of the complete genome of HBV genotypes F and H found in Brazil and Mexico using the next generation sequencing method.Ann Hepatol. 2022 Jan;27 Suppl 1:100569. doi: 10.1016/j.aohep.2021.100569. Epub 2021 Oct 29. Ann Hepatol. 2022. PMID: 34757035
-
Hepatitis B virus genotypes: comparison of genotyping methods.Rev Med Virol. 2004 Jan-Feb;14(1):3-16. doi: 10.1002/rmv.400. Rev Med Virol. 2004. PMID: 14716688 Review.
-
Next-generation sequencing for the diagnosis of hepatitis B: current status and future prospects.Expert Rev Mol Diagn. 2021 Apr;21(4):381-396. doi: 10.1080/14737159.2021.1913055. Epub 2021 Apr 21. Expert Rev Mol Diagn. 2021. PMID: 33880971 Review.
Cited by
-
An Oxford Nanopore Technology-Based Hepatitis B Virus Sequencing Protocol Suitable for Genomic Surveillance Within Clinical Diagnostic Settings.Int J Mol Sci. 2024 Oct 31;25(21):11702. doi: 10.3390/ijms252111702. Int J Mol Sci. 2024. PMID: 39519254 Free PMC article.
References
-
- World Health Organization Hepatitis B. [(accessed on 16 May 2023)]; Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

