Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Hydrolyze Recombinant RBD and Nucleocapsid (N) Proteins
- PMID: 38790969
- PMCID: PMC11118737
- DOI: 10.3390/biomedicines12051007
Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Hydrolyze Recombinant RBD and Nucleocapsid (N) Proteins
Abstract
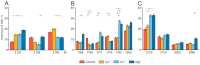
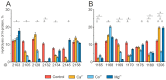
Antibodies are protein molecules whose primary function is to recognize antigens. However, recent studies have demonstrated their ability to hydrolyze specific substrates, such as proteins, oligopeptides, and nucleic acids. In 2023, two separate teams of researchers demonstrated the proteolytic activity of natural plasma antibodies from COVID-19 convalescents. These antibodies were found to hydrolyze the S-protein and corresponding oligopeptides. Our study shows that for antibodies with affinity to recombinant structural proteins of the SARS-CoV-2: S-protein, its fragment RBD and N-protein can only hydrolyze the corresponding protein substrates and are not cross-reactive. By using strict criteria, we have confirmed that this proteolytic activity is an intrinsic property of antibodies and is not caused by impurities co-eluting with them. This discovery suggests that natural proteolytic antibodies that hydrolyze proteins of the SARS-CoV-2 virus may have a positive impact on disease pathogenesis. It is also possible for these antibodies to work in combination with other antibodies that bind specific epitopes to enhance the process of virus neutralization.
Keywords: COVID-19; IgG; N-protein; RBD; SARS-CoV-2; autoimmunity; catalytic antibodies; coronavirus; proteolytic antibodies.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analysis, or interpretation of the data, in the writing of the manuscript, or in the decision to publish the results.
Figures
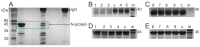
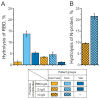
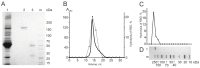
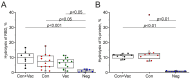
Similar articles
-
Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Recognize and Hydrolyze Oligopeptides Corresponding to the S-Protein of SARS-CoV-2.Vaccines (Basel). 2023 Sep 15;11(9):1494. doi: 10.3390/vaccines11091494. Vaccines (Basel). 2023. PMID: 37766170 Free PMC article.
-
Dissection of Antibody Responses of Gam-COVID-Vac-Vaccinated Subjects Suggests Involvement of Epitopes Outside RBD in SARS-CoV-2 Neutralization.Int J Mol Sci. 2023 Mar 7;24(6):5104. doi: 10.3390/ijms24065104. Int J Mol Sci. 2023. PMID: 36982183 Free PMC article.
-
Cross-Reactivity of SARS-CoV-2 Nucleocapsid-Binding Antibodies and Its Implication for COVID-19 Serology Tests.Viruses. 2022 Sep 14;14(9):2041. doi: 10.3390/v14092041. Viruses. 2022. PMID: 36146847 Free PMC article.
-
Heterogeneous antibodies against SARS-CoV-2 spike receptor binding domain and nucleocapsid with implications for COVID-19 immunity.JCI Insight. 2020 Sep 17;5(18):e142386. doi: 10.1172/jci.insight.142386. JCI Insight. 2020. PMID: 32796155 Free PMC article.
-
[Investigation of SARS-CoV-2-Specific Humoral and Cellular Immunity Values in Health Care Workers with COVID-19 Disease and Administered with COVID-19 Vaccine].Mikrobiyol Bul. 2022 Jul;56(3):480-492. doi: 10.5578/mb.20229708. Mikrobiyol Bul. 2022. PMID: 35960239 Turkish.
Cited by
-
Circulating miRNAs in the Plasma of Post-COVID-19 Patients with Typical Recovery and Those with Long-COVID Symptoms: Regulation of Immune Response-Associated Pathways.Noncoding RNA. 2024 Sep 2;10(5):48. doi: 10.3390/ncrna10050048. Noncoding RNA. 2024. PMID: 39311385 Free PMC article.
References
-
- Lerner R.A., Tramontano A. Antibodies as Enzymes. Trends Biochem. Sci. 1987;12:427–430. doi: 10.1016/0968-0004(87)90208-8. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

