Unravelling the Role of Candida albicans Prn1 in the Oxidative Stress Response through a Proteomics Approach
- PMID: 38790632
- PMCID: PMC11118716
- DOI: 10.3390/antiox13050527
Unravelling the Role of Candida albicans Prn1 in the Oxidative Stress Response through a Proteomics Approach
Abstract
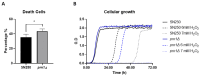
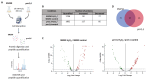
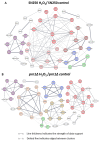
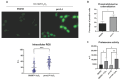
Candida albicans Prn1 is a protein with an unknown function similar to mammalian Pirin. It also has orthologues in other pathogenic fungi, but not in Saccharomyces cerevisiae. Prn1 highly increases its abundance in response to H2O2 treatment; thus, to study its involvement in the oxidative stress response, a C. albicans prn1∆ mutant and the corresponding wild-type strain SN250 have been studied. Under H2O2 treatment, Prn1 absence led to a higher level of reactive oxygen species (ROS) and a lower survival rate, with a higher percentage of death by apoptosis, confirming its relevant role in oxidative detoxication. The quantitative differential proteomics studies of both strains in the presence and absence of H2O2 indicated a lower increase in proteins with oxidoreductase activity after the treatment in the prn1∆ strain, as well as an increase in proteasome-activating proteins, corroborated by in vivo measurements of proteasome activity, with respect to the wild type. In addition, remarkable differences in the abundance of some transcription factors were observed between mutant and wild-type strains, e.g., Mnl1 or Nrg1, an Mnl1 antagonist. orf19.4850, a protein orthologue to S. cerevisiae Cub1, has shown its involvement in the response to H2O2 and in proteasome function when Prn1 is highly expressed in the wild type.
Keywords: C. albicans; Cub1; Mnl1; Nrg1; Pirin; Prn1; apoptosis; mitochondria; oxidative stress response; proteasome; proteomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Extending the Proteomic Characterization of Candida albicans Exposed to Stress and Apoptotic Inducers through Data-Independent Acquisition Mass Spectrometry.mSystems. 2021 Oct 26;6(5):e0094621. doi: 10.1128/mSystems.00946-21. Epub 2021 Oct 5. mSystems. 2021. PMID: 34609160 Free PMC article.
-
MNL1 regulates weak acid-induced stress responses of the fungal pathogen Candida albicans.Mol Biol Cell. 2008 Oct;19(10):4393-403. doi: 10.1091/mbc.e07-09-0946. Epub 2008 Jul 23. Mol Biol Cell. 2008. PMID: 18653474 Free PMC article.
-
Systematic identification and characterization of five transcription factors mediating the oxidative stress response in Candida albicans.Microb Pathog. 2024 Feb;187:106507. doi: 10.1016/j.micpath.2023.106507. Epub 2023 Dec 23. Microb Pathog. 2024. PMID: 38145792
-
Candida and candidaemia. Susceptibility and epidemiology.Dan Med J. 2013 Nov;60(11):B4698. Dan Med J. 2013. PMID: 24192246 Review.
-
Apoptotic Factors, CaNma111 and CaYbh3, Function in Candida albicans Filamentation by Regulating the Hyphal Suppressors, Nrg1 and Tup1.J Microbiol. 2023 Apr;61(4):403-409. doi: 10.1007/s12275-023-00034-8. Epub 2023 Mar 27. J Microbiol. 2023. PMID: 36972003 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

