The Landscape of Presence/Absence Variations during the Improvement of Rice
- PMID: 38790274
- PMCID: PMC11120952
- DOI: 10.3390/genes15050645
The Landscape of Presence/Absence Variations during the Improvement of Rice
Abstract
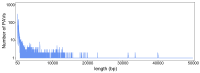
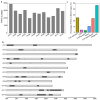
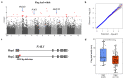
Rice is one of the most important staple crops in the world; therefore, the improvement of rice holds great significance for enhancing agricultural production and addressing food security challenges. Although there have been numerous studies on the role of single-nucleotide polymorphisms (SNPs) in rice improvement with the development of next-generation sequencing technologies, research on the role of presence/absence variations (PAVs) in the improvement of rice is limited. In particular, there is a scarcity of studies exploring the traits and genes that may be affected by PAVs in rice. Here, we extracted PAVs utilizing resequencing data from 148 improved rice varieties distributed in Asia. We detected a total of 33,220 PAVs and found that the number of variations decreased gradually as the length of the PAVs increased. The number of PAVs was the highest on chromosome 1. Furthermore, we identified a 6 Mb hotspot region on chromosome 11 containing 1091 PAVs in which there were 29 genes related to defense responses. By conducting a genome-wide association study (GWAS) using PAV variation data and phenotypic data for five traits (flowering time, plant height, flag leaf length, flag leaf width, and panicle number) across all materials, we identified 186 significantly associated PAVs involving 20 cloned genes. A haplotype analysis and expression analysis of candidate genes revealed that important genes might be affected by PAVs, such as the flowering time gene OsSFL1 and the flag leaf width gene NAL1. Our work investigated the pattern in PAVs and explored important PAV key functional genes associated with agronomic traits. Consequently, these results provide potential and exploitable genetic resources for rice breeding.
Keywords: genome-wide association study; presence/absence variations; rice improvement.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Genetic variation and association mapping for 12 agronomic traits in indica rice.BMC Genomics. 2015 Dec 16;16:1067. doi: 10.1186/s12864-015-2245-2. BMC Genomics. 2015. PMID: 26673149 Free PMC article.
-
Genome-Wide Association Mapping for Yield and Yield-Related Traits in Rice (Oryza Sativa L.) Using SNPs Markers.Genes (Basel). 2023 May 15;14(5):1089. doi: 10.3390/genes14051089. Genes (Basel). 2023. PMID: 37239449 Free PMC article.
-
Genome-Wide Association Study of Root System Development at Seedling Stage in Rice.Genes (Basel). 2020 Nov 25;11(12):1395. doi: 10.3390/genes11121395. Genes (Basel). 2020. PMID: 33255557 Free PMC article.
-
A Rice Genetic Improvement Boom by Next Generation Sequencing.Curr Issues Mol Biol. 2018;27:109-126. doi: 10.21775/cimb.027.109. Epub 2017 Sep 8. Curr Issues Mol Biol. 2018. PMID: 28885178 Review.
-
Advances in genome-wide association studies of complex traits in rice.Theor Appl Genet. 2020 May;133(5):1415-1425. doi: 10.1007/s00122-019-03473-3. Epub 2019 Nov 12. Theor Appl Genet. 2020. PMID: 31720701 Review.
References
-
- Wang X.Q., Wang W.S., Tai S.S., Li M., Gao Q., Hu Z.Q., Hu W.S., Wu Z.C., Zhu X.Y., Xie J.Y., et al. Selective and comparative genome architecture of Asian cultivated rice (Oryza sativa L.) attributed to domestication and modern breeding. J. Adv. Res. 2022;42:1–16. doi: 10.1016/j.jare.2022.08.004. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

