In Silico Analysis of Protein-Protein Interactions of Putative Endoplasmic Reticulum Metallopeptidase 1 in Schizosaccharomyces pombe
- PMID: 38785548
- PMCID: PMC11120530
- DOI: 10.3390/cimb46050280
In Silico Analysis of Protein-Protein Interactions of Putative Endoplasmic Reticulum Metallopeptidase 1 in Schizosaccharomyces pombe
Abstract
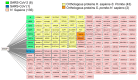
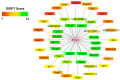
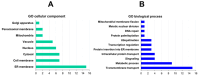
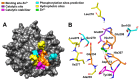
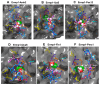
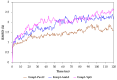
Ermp1 is a putative metalloprotease from Schizosaccharomyces pombe and a member of the Fxna peptidases. Although their function is unknown, orthologous proteins from rats and humans have been associated with the maturation of ovarian follicles and increased ER stress. This study focuses on proposing the first prediction of PPI by comparison of the interologues between humans and yeasts, as well as the molecular docking and dynamics of the M28 domain of Ermp1 with possible target proteins. As results, 45 proteins are proposed that could interact with the metalloprotease. Most of these proteins are related to the transport of Ca2+ and the metabolism of amino acids and proteins. Docking and molecular dynamics suggest that the M28 domain of Ermp1 could hydrolyze leucine and methionine residues of Amk2, Ypt5 and Pex12. These results could support future experimental investigations of other Fxna peptidases, such as human ERMP1.
Keywords: Ermp1; S. pombe; metalloprotease; molecular docking; molecular dynamic; protein–protein interaction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Fxna, a novel gene differentially expressed in the rat ovary at the time of folliculogenesis, is required for normal ovarian histogenesis.Development. 2007 Mar;134(5):945-57. doi: 10.1242/dev.02795. Epub 2007 Jan 31. Development. 2007. PMID: 17267443
-
miR-9-5p attenuates ischemic stroke through targeting ERMP1-mediated endoplasmic reticulum stress.Acta Histochem. 2019 Nov;121(8):151438. doi: 10.1016/j.acthis.2019.08.005. Epub 2019 Sep 7. Acta Histochem. 2019. PMID: 31500865
-
ERMP1, a novel potential oncogene involved in UPR and oxidative stress defense, is highly expressed in human cancer.Oncotarget. 2016 Sep 27;7(39):63596-63610. doi: 10.18632/oncotarget.11550. Oncotarget. 2016. PMID: 27566589 Free PMC article.
-
Vesicle-mediated protein transport pathways to the vacuole in Schizosaccharomyces pombe.Cell Struct Funct. 2003 Oct;28(5):399-417. doi: 10.1247/csf.28.399. Cell Struct Funct. 2003. PMID: 14745133 Review.
-
Identification and phylogenetic classification of eleven putative P-type calcium transport ATPase genes in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe.Biosci Rep. 1996 Apr;16(2):75-85. doi: 10.1007/BF01206198. Biosci Rep. 1996. PMID: 8790914 Review.
References
-
- Grandi A., Santi A., Campagnoli S., Parri M., De Camilli E., Song C., Jin B., Lacombe A., Castori-Eppenberger S., Sarmientos P., et al. ERMP1, a Novel Potential Oncogene Involved in UPR and Oxidative Stress Defense, Is Highly Expressed in Human Cancer. Oncotarget. 2016;7:63596–63610. doi: 10.18632/oncotarget.11550. - DOI - PMC - PubMed
-
- Garcia-Rudaz C., Luna F., Tapia V., Kerr B., Colgin L., Galimi F., Dissen G.A., Rawlings N.D., Ojeda S.R. Fxna, a Novel Gene Differentially Expressed in the Rat Ovary at the Time of Folliculogenesis, Is Required for Normal Ovarian Histogenesis. Development. 2007;134:945–957. doi: 10.1242/dev.02795. - DOI - PubMed
-
- Dastghaib S., Mokarram P., Erfani M., Ghavami S., Hosseini S.V., Zamani M. Endoplasmic Reticulum Metallo Protease 1, a Triggering Factor for Unfolded Protein Response and Promising Target in Colorectal Cancer. Biologia. 2021;76:2403–2411. doi: 10.1007/s11756-021-00769-y. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

