Exploring gene regulatory interaction networks and predicting therapeutic molecules for hypopharyngeal cancer and EGFR-mutated lung adenocarcinoma
- PMID: 38783639
- PMCID: PMC11216941
- DOI: 10.1002/2211-5463.13807
Exploring gene regulatory interaction networks and predicting therapeutic molecules for hypopharyngeal cancer and EGFR-mutated lung adenocarcinoma
Abstract
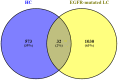
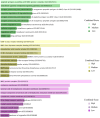
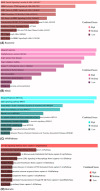
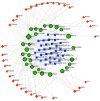
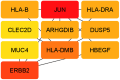
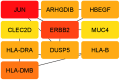
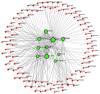
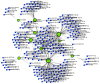
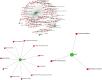
Hypopharyngeal cancer is a disease that is associated with EGFR-mutated lung adenocarcinoma. Here we utilized a bioinformatics approach to identify genetic commonalities between these two diseases. To this end, we examined microarray datasets from GEO (Gene Expression Omnibus) to identify differentially expressed genes, common genes, and hub genes between the selected two diseases. Our analyses identified potential therapeutic molecules for the selected diseases based on 10 hub genes with the highest interactions according to the degree topology method and the maximum clique centrality (MCC). These therapeutic molecules may have the potential for simultaneous treatment of these diseases.
Keywords: EGFR‐mutated lung adenocarcinoma; Gene Expression Omnibus; degree topology, therapeutic molecule; hub gene; hypopharyngeal cancer.
© 2024 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Decreased HLF Expression Predicts Poor Survival in Lung Adenocarcinoma.Med Sci Monit. 2021 May 12;27:e929333. doi: 10.12659/MSM.929333. Med Sci Monit. 2021. PMID: 33979320 Free PMC article.
-
Identification of lung adenocarcinoma biomarkers based on bioinformatic analysis and human samples.Oncol Rep. 2020 May;43(5):1437-1450. doi: 10.3892/or.2020.7526. Epub 2020 Feb 28. Oncol Rep. 2020. PMID: 32323809 Free PMC article.
-
Identification of differential protein-coding gene expressions in early phase lung adenocarcinoma.Thorac Cancer. 2018 Feb;9(2):234-240. doi: 10.1111/1759-7714.12569. Epub 2017 Dec 20. Thorac Cancer. 2018. PMID: 29266838 Free PMC article.
-
Microarray analysis of the expression profile of immune-related gene in rapid recurrence early-stage lung adenocarcinoma.J Cancer Res Clin Oncol. 2020 Sep;146(9):2299-2310. doi: 10.1007/s00432-020-03287-7. Epub 2020 Jun 18. J Cancer Res Clin Oncol. 2020. PMID: 32556504 Free PMC article.
-
Role of microRNAs in epidermal growth factor receptor signaling pathway in cervical cancer.Mol Biol Rep. 2020 Jun;47(6):4553-4568. doi: 10.1007/s11033-020-05494-4. Epub 2020 May 7. Mol Biol Rep. 2020. PMID: 32383136 Review.
Cited by
-
Toward reliable diabetes prediction: Innovations in data engineering and machine learning applications.Digit Health. 2024 Aug 21;10:20552076241271867. doi: 10.1177/20552076241271867. eCollection 2024 Jan-Dec. Digit Health. 2024. PMID: 39175924 Free PMC article.
References
-
- Phoebe Chen Y‐P and Chen F (2008) Identifying targets for drug discovery using bioinformatics. Expert Opin Ther Targets 12, 383–389. - PubMed
-
- Hanif MA (2018) Head and neck cancer‐Bangladesh perspective. Bangladesh J Otorhin 24, 1–2.
-
- Sanders O and Pathak S (2022) Hypopharyngeal cancer. - PubMed
-
- Hoffmann M, Görögh T, Gottschlich S, Lohrey C, Rittgen W, Ambrosch P, Schwarz E and Kahn T (2005) Human Papillomaviruses in head and neck cancer: 8 year‐survival‐analysis of 73 patients. Cancer Lett 218, 199–206. - PubMed
-
- Rodrigo JP, Hermsen MA, Fresno MF, Brakenhoff RH, García‐Velasco F, Snijders PJ, Heideman DA and García‐Pedrero JM (2015) Prevalence of human papillomavirus in laryngeal and hypopharyngeal squamous cell carcinomas in northern Spain. Cancer Epidemiol 39, 37–41. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

