Potential involvement of beta-lactamase homologous proteins in resistance to beta-lactam antibiotics in gram-negative bacteria of the ESKAPEE group
- PMID: 38778284
- PMCID: PMC11112869
- DOI: 10.1186/s12864-024-10410-2
Potential involvement of beta-lactamase homologous proteins in resistance to beta-lactam antibiotics in gram-negative bacteria of the ESKAPEE group
Abstract
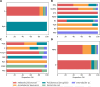
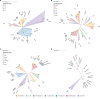
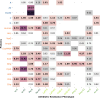
Enzymatic degradation mediated by beta-lactamases constitutes one of the primary mechanisms of resistance to beta-lactam antibiotics in gram-negative bacteria. This enzyme family comprises four molecular classes, categorized into serine beta-lactamases (Classes A, C, and D) and zinc-dependent metallo-beta-lactamases (Class B). Gram-negative bacteria producing beta-lactamase are of significant concern, particularly due to their prevalence in nosocomial infections. A comprehensive understanding of the evolution and dissemination of this enzyme family is essential for effective control of these pathogens. In this study, we conducted the prospecting, phylogenetic analysis, and in silico analysis of beta-lactamases and homologous proteins identified in 1827 bacterial genomes with phenotypic data on beta-lactam resistance. These genomes were distributed among Klebsiella pneumoniae (45%), Acinetobacter baumannii (31%), Pseudomonas aeruginosa (14%), Escherichia coli (6%), and Enterobacter spp. (4%). Using an HMM profile and searching for conserved domains, we mined 2514, 8733, 5424, and 2957 proteins for molecular classes A, B, C, and D, respectively. This set of proteins encompasses canonical subfamilies of beta-lactamases as well as hypothetical proteins and other functional groups. Canonical beta-lactamases were found to be phylogenetically distant from hypothetical proteins, which, in turn, are closer to other representatives of the penicillin-binding-protein (PBP-like) and metallo-beta-lactamase (MBL) families. The catalytic amino acid residues characteristic of beta-lactamases were identified from the sequence alignment and revealed that motifs are less conserved in homologous groups than in beta-lactamases. After comparing the frequency of protein groups in genomes of resistant strains with those of sensitive ones applying Fisher's exact test and relative risk, it was observed that some groups of homologous proteins to classes B and C are more common in the genomes of resistant strains, particularly to carbapenems. We identified the beta-lactamase-like domain widely distributed in gram-negative species of the ESKAPEE group, which highlights its importance in the context of beta-lactam resistance. Some hypothetical homologous proteins have been shown to potentially possess promiscuous activity against beta-lactam antibiotics, however, they do not appear to expressly determine the resistance phenotype. The selective pressure due to the widespread use of antibiotics may favor the optimization of these functions for specialized resistance enzymes.
Keywords: Beta-lactamase; ESKAPEE; beta-lactam antibiotics; homologous protein; resistance.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
β-Lactamases and β-Lactamase Inhibitors in the 21st Century.J Mol Biol. 2019 Aug 23;431(18):3472-3500. doi: 10.1016/j.jmb.2019.04.002. Epub 2019 Apr 5. J Mol Biol. 2019. PMID: 30959050 Free PMC article. Review.
-
Cyclic Boronates Inhibit All Classes of β-Lactamases.Antimicrob Agents Chemother. 2017 Mar 24;61(4):e02260-16. doi: 10.1128/AAC.02260-16. Print 2017 Apr. Antimicrob Agents Chemother. 2017. PMID: 28115348 Free PMC article.
-
[Antimicrobial susceptibility and molecular mechanisms of resistance to beta-lactams of gram-negative microorganisms--causative agents of nosocomial infections].Zh Mikrobiol Epidemiol Immunobiol. 2007 Sep-Oct;(5):16-20. Zh Mikrobiol Epidemiol Immunobiol. 2007. PMID: 18038541 Russian.
-
The emergence and implications of metallo-beta-lactamases in Gram-negative bacteria.Clin Microbiol Infect. 2005 Nov;11 Suppl 6:2-9. doi: 10.1111/j.1469-0691.2005.01264.x. Clin Microbiol Infect. 2005. PMID: 16209700 Review.
-
Metallo-β-lactamases: a last frontier for β-lactams?Lancet Infect Dis. 2011 May;11(5):381-93. doi: 10.1016/S1473-3099(11)70056-1. Lancet Infect Dis. 2011. PMID: 21530894 Review.
References
-
- World Health Organization (WHO) Antimicrobial resistance: global report on surveillance. World Health Organization: Geneva, Switzerland; 2014.
-
- Ngoi ST, Chong CW, Ponnampalavanar SSLS, Tang SN, Idris N, Abdul Jabar K, et al. Genetic mechanisms and correlated risk factors of antimicrobial-resistant ESKAPEE pathogens isolated in a tertiary hospital in Malaysia. Antimicrob Resist Infect Control. 2021;10(1):70. doi: 10.1186/s13756-021-00936-5. - DOI - PMC - PubMed
-
- World Health Organization (WHO), editor. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. Geneva, Switzerland: World Health Organization; 2017.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

