Acquisition of epithelial plasticity in human chronic liver disease
- PMID: 38778114
- PMCID: PMC11153150
- DOI: 10.1038/s41586-024-07465-2
Acquisition of epithelial plasticity in human chronic liver disease
Abstract
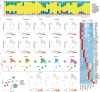
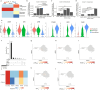
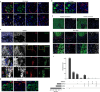
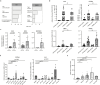
For many adult human organs, tissue regeneration during chronic disease remains a controversial subject. Regenerative processes are easily observed in animal models, and their underlying mechanisms are becoming well characterized1-4, but technical challenges and ethical aspects are limiting the validation of these results in humans. We decided to address this difficulty with respect to the liver. This organ displays the remarkable ability to regenerate after acute injury, although liver regeneration in the context of recurring injury remains to be fully demonstrated. Here we performed single-nucleus RNA sequencing (snRNA-seq) on 47 liver biopsies from patients with different stages of metabolic dysfunction-associated steatotic liver disease to establish a cellular map of the liver during disease progression. We then combined these single-cell-level data with advanced 3D imaging to reveal profound changes in the liver architecture. Hepatocytes lose their zonation and considerable reorganization of the biliary tree takes place. More importantly, our study uncovers transdifferentiation events that occur between hepatocytes and cholangiocytes without the presence of adult stem cells or developmental progenitor activation. Detailed analyses and functional validations using cholangiocyte organoids confirm the importance of the PI3K-AKT-mTOR pathway in this process, thereby connecting this acquisition of plasticity to insulin signalling. Together, our data indicate that chronic injury creates an environment that induces cellular plasticity in human organs, and understanding the underlying mechanisms of this process could open new therapeutic avenues in the management of chronic diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

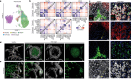
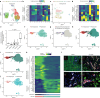
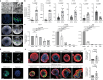

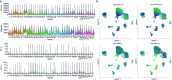




Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Dynamic Field Theory of Executive Function: Identifying Early Neurocognitive Markers.Monogr Soc Res Child Dev. 2024 Dec;89(3):7-109. doi: 10.1111/mono.12478. Monogr Soc Res Child Dev. 2024. PMID: 39628288 Free PMC article.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
DNA methylome analysis reveals epigenetic alteration of complement genes in advanced metabolic dysfunction-associated steatotic liver disease.Clin Mol Hepatol. 2024 Oct;30(4):824-844. doi: 10.3350/cmh.2024.0229. Epub 2024 Jul 25. Clin Mol Hepatol. 2024. PMID: 39048522 Free PMC article.
-
Targeting Liver Epsins Ameliorates Dyslipidemia in Atherosclerosis.bioRxiv [Preprint]. 2024 Aug 27:2024.08.26.609742. doi: 10.1101/2024.08.26.609742. bioRxiv. 2024. PMID: 39253478 Free PMC article. Preprint.
-
Transcriptome Analyses of Liver Sinusoidal Endothelial Cells Reveal a Consistent List of Candidate Genes Associated with Endothelial Dysfunction and the Fibrosis Progression.Curr Issues Mol Biol. 2024 Jul 25;46(8):7997-8014. doi: 10.3390/cimb46080473. Curr Issues Mol Biol. 2024. PMID: 39194690 Free PMC article.
-
Liver regeneration after injury: Mechanisms, cellular interactions and therapeutic innovations.Clin Transl Med. 2024 Aug;14(8):e1812. doi: 10.1002/ctm2.1812. Clin Transl Med. 2024. PMID: 39152680 Free PMC article. Review.
-
Using different zebrafish models to explore liver regeneration.Front Cell Dev Biol. 2024 Oct 31;12:1485773. doi: 10.3389/fcell.2024.1485773. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39544362 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

