Role of N-linked glycosylation in porcine reproductive and respiratory syndrome virus (PRRSV) infection
- PMID: 38776134
- PMCID: PMC11165596
- DOI: 10.1099/jgv.0.001994
Role of N-linked glycosylation in porcine reproductive and respiratory syndrome virus (PRRSV) infection
Abstract
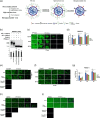
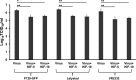
Porcine reproductive and respiratory syndrome (PRRSV) is an enveloped single-stranded positive-sense RNA virus and one of the main pathogens that causes the most significant economical losses in the swine-producing countries. PRRSV is currently divided into two distinct species, PRRSV-1 and PRRSV-2. The PRRSV virion envelope is composed of four glycosylated membrane proteins and three non-glycosylated envelope proteins. Previous work has suggested that PRRSV-linked glycans are critical structural components for virus assembly. In addition, it has been proposed that PRRSV glycans are implicated in the interaction with host cells and critical for virus infection. In contrast, recent findings showed that removal of N-glycans from PRRSV does not influence virus infection of permissive cells. Thus, there are not sufficient evidences to indicate compellingly that N-glycans present in the PRRSV envelope play a direct function in viral infection. To gain insights into the role of N-glycosylation in PRRSV infection, we analysed the specific contribution of the envelope protein-linked N-glycans to infection of permissive cells. For this purpose, we used a novel strategy to modify envelope protein-linked N-glycans that consists of production of monoglycosylated PRRSV and viral glycoproteins with different glycan states. Our results showed that removal or alteration of N-glycans from PRRSV affected virus infection. Specifically, we found that complex N-glycans are required for an efficient infection in cell cultures. Furthermore, we found that presence of high mannose type glycans on PRRSV surface is the minimal requirement for a productive viral infection. Our findings also show that PRRSV-1 and PRRSV-2 have different requirements of N-glycan structure for an optimal infection. In addition, we demonstrated that removal of N-glycans from PRRSV does not affect viral attachment, suggesting that these carbohydrates played a major role in regulating viral entry. In agreement with these findings, by performing immunoprecipitation assays and colocalization experiments, we found that N-glycans present in the viral envelope glycoproteins are not required to bind to the essential viral receptor CD163. Finally, we found that the presence of N-glycans in CD163 is not required for PRRSV infection.
Keywords: N-glycosylation; PRRSV; viral envelope glycoproteins; viral infectivity.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


Similar articles
-
Influence of N-linked glycosylation of minor proteins of porcine reproductive and respiratory syndrome virus on infectious virus recovery and receptor interaction.Virology. 2012 Jul 20;429(1):1-11. doi: 10.1016/j.virol.2012.03.010. Epub 2012 Apr 25. Virology. 2012. PMID: 22537809
-
Functional analysis of porcine reproductive and respiratory syndrome virus N-glycans in infection of permissive cells.Virology. 2015 Mar;477:82-88. doi: 10.1016/j.virol.2015.01.005. Epub 2015 Feb 6. Virology. 2015. PMID: 25662311
-
Glycosyl-phosphatidylinositol (GPI)-anchored membrane association of the porcine reproductive and respiratory syndrome virus GP4 glycoprotein and its co-localization with CD163 in lipid rafts.Virology. 2012 Mar 1;424(1):18-32. doi: 10.1016/j.virol.2011.12.009. Epub 2012 Jan 4. Virology. 2012. PMID: 22222209 Free PMC article.
-
PRRS virus receptors and their role for pathogenesis.Vet Microbiol. 2015 Jun 12;177(3-4):229-41. doi: 10.1016/j.vetmic.2015.04.002. Epub 2015 Apr 13. Vet Microbiol. 2015. PMID: 25912022 Review.
-
Gene editing as applied to prevention of reproductive porcine reproductive and respiratory syndrome.Mol Reprod Dev. 2017 Sep;84(9):926-933. doi: 10.1002/mrd.22811. Epub 2017 Jun 8. Mol Reprod Dev. 2017. PMID: 28390179 Review.
Cited by
-
A Comprehensive Review on Porcine Reproductive and Respiratory Syndrome Virus with Emphasis on Immunity.Vaccines (Basel). 2024 Aug 22;12(8):942. doi: 10.3390/vaccines12080942. Vaccines (Basel). 2024. PMID: 39204065 Free PMC article. Review.
References
-
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. - PubMed
-
- Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, Harrison RL, et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017) Arch Virol. 2017;162:2505–2538. doi: 10.1007/s00705-017-3358-5. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

