Differential 3D genome architecture and imprinted gene expression: cause or consequence?
- PMID: 38775198
- PMCID: PMC11346452
- DOI: 10.1042/BST20230143
Differential 3D genome architecture and imprinted gene expression: cause or consequence?
Abstract
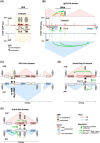
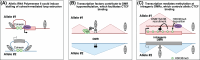
Imprinted genes provide an attractive paradigm to unravel links between transcription and genome architecture. The parental allele-specific expression of these essential genes - which are clustered in chromosomal domains - is mediated by parental methylation imprints at key regulatory DNA sequences. Recent chromatin conformation capture (3C)-based studies show differential organization of topologically associating domains between the parental chromosomes at imprinted domains, in embryonic stem and differentiated cells. At several imprinted domains, differentially methylated regions show allelic binding of the insulator protein CTCF, and linked focal retention of cohesin, at the non-methylated allele only. This generates differential patterns of chromatin looping between the parental chromosomes, already in the early embryo, and thereby facilitates the allelic gene expression. Recent research evokes also the opposite scenario, in which allelic transcription contributes to the differential genome organization, similarly as reported for imprinted X chromosome inactivation. This may occur through epigenetic effects on CTCF binding, through structural effects of RNA Polymerase II, or through imprinted long non-coding RNAs that have chromatin repressive functions. The emerging picture is that epigenetically-controlled differential genome architecture precedes and facilitates imprinted gene expression during development, and that at some domains, conversely, the mono-allelic gene expression also influences genome architecture.
Keywords: CTCF; chromatin architecture; chromatin loop; genomic imprinting; topologically associating domain.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


Similar articles
-
CTCF modulates allele-specific sub-TAD organization and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains.Genome Biol. 2019 Dec 12;20(1):272. doi: 10.1186/s13059-019-1896-8. Genome Biol. 2019. PMID: 31831055 Free PMC article.
-
Differential 3D chromatin organization and gene activity in genomic imprinting.Curr Opin Genet Dev. 2020 Apr;61:17-24. doi: 10.1016/j.gde.2020.03.004. Epub 2020 Apr 13. Curr Opin Genet Dev. 2020. PMID: 32299027 Review.
-
Genome-wide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions.Genome Res. 2013 Oct;23(10):1624-35. doi: 10.1101/gr.150136.112. Epub 2013 Jun 26. Genome Res. 2013. PMID: 23804403 Free PMC article.
-
Exploring chromatin structural roles of non-coding RNAs at imprinted domains.Biochem Soc Trans. 2021 Aug 27;49(4):1867-1879. doi: 10.1042/BST20210758. Biochem Soc Trans. 2021. PMID: 34338292 Free PMC article. Review.
-
Impact of 3D genome organization, guided by cohesin and CTCF looping, on sex-biased chromatin interactions and gene expression in mouse liver.Epigenetics Chromatin. 2020 Jul 17;13(1):30. doi: 10.1186/s13072-020-00350-y. Epigenetics Chromatin. 2020. PMID: 32680543 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

