Modeling reveals the strength of weak interactions in stacked-ring assembly
- PMID: 38762753
- PMCID: PMC11267433
- DOI: 10.1016/j.bpj.2024.05.015
Modeling reveals the strength of weak interactions in stacked-ring assembly
Abstract
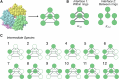
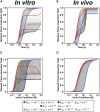
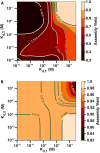
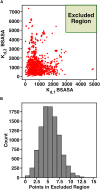
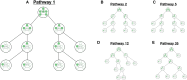
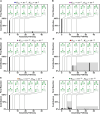
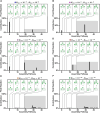
Cells employ many large macromolecular machines for the execution and regulation of processes that are vital for cell and organismal viability. Interestingly, cells cannot synthesize these machines as functioning units. Instead, cells synthesize the molecular parts that must then assemble into the functional complex. Many important machines, including chaperones such as GroEL and proteases such as the proteasome, comprise protein rings that are stacked on top of one another. While there is some experimental data regarding how stacked-ring complexes such as the proteasome self-assemble, a comprehensive understanding of the dynamics of stacked-ring assembly is currently lacking. Here, we developed a mathematical model of stacked-trimer assembly and performed an analysis of the assembly of the stacked homomeric trimer, which is the simplest stacked-ring architecture. We found that stacked rings are particularly susceptible to a form of kinetic trapping that we term "deadlock," in which the system gets stuck in a state where there are many large intermediates that are not the fully assembled structure but that cannot productively react. When interaction affinities are uniformly strong, deadlock severely limits assembly yield. We thus predicted that stacked rings would avoid situations where all interfaces in the structure have high affinity. Analysis of available crystal structures indicated that indeed the majority-if not all-of stacked trimers do not contain uniformly strong interactions. Finally, to better understand the origins of deadlock, we developed a formal pathway analysis and showed that, when all the binding affinities are strong, many of the possible pathways are utilized. In contrast, optimal assembly strategies utilize only a small number of pathways. Our work suggests that deadlock is a critical factor influencing the evolution of macromolecular machines and provides general principles for understanding the self-assembly efficiency of existing machines.
Copyright © 2024 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
Similar articles
-
Optimizing ring assembly reveals the strength of weak interactions.Proc Natl Acad Sci U S A. 2012 Feb 14;109(7):2348-53. doi: 10.1073/pnas.1113095109. Epub 2012 Jan 30. Proc Natl Acad Sci U S A. 2012. PMID: 22308356 Free PMC article.
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
Homomeric ring assemblies of eukaryotic Sm proteins have affinity for both RNA and DNA. Crystal structure of an oligomeric complex of yeast SmF.J Biol Chem. 2003 May 9;278(19):17291-8. doi: 10.1074/jbc.M211826200. Epub 2003 Mar 4. J Biol Chem. 2003. PMID: 12618433
-
Structure, dynamics, assembly, and evolution of protein complexes.Annu Rev Biochem. 2015;84:551-75. doi: 10.1146/annurev-biochem-060614-034142. Epub 2014 Dec 8. Annu Rev Biochem. 2015. PMID: 25494300 Review.
-
Assembly of the 20S proteasome.Biochim Biophys Acta. 2014 Jan;1843(1):2-12. doi: 10.1016/j.bbamcr.2013.03.008. Epub 2013 Mar 16. Biochim Biophys Acta. 2014. PMID: 23507199 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

