NUP43 promotes PD-L1/nPD-L1/PD-L1 feedback loop via TM4SF1/JAK/STAT3 pathway in colorectal cancer progression and metastatsis
- PMID: 38762481
- PMCID: PMC11102480
- DOI: 10.1038/s41420-024-02025-z
NUP43 promotes PD-L1/nPD-L1/PD-L1 feedback loop via TM4SF1/JAK/STAT3 pathway in colorectal cancer progression and metastatsis
Abstract
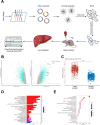
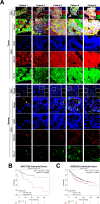
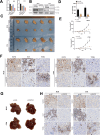
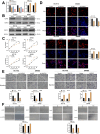
Programmed cell death-ligand 1 (PD-L1) has a significant role in tumor progression and metastasis, facilitating tumor cell evasion from immune surveillance. PD-L1 can be detected in the tumor cell nucleus and exert an oncogenic effect by nuclear translocation. Colorectal cancer (CRC) progression and liver metastasis (CCLM) are among the most lethal diseases worldwide, but the mechanism of PD-L1 nuclear translocation in CRC and CCLM remains to be fully understood. In this study, using CRISPR-Cas9-based genome-wide screening combined with RNA-seq, we found that the oncogenic factor NUP43 impacted the process of PD-L1 nuclear translocation by regulating the expression level of the PD-L1 chaperone protein IPO5. Subsequent investigation revealed that this process could stimulate the expression of tumor-promoting factor TM4SF1 and further activate the JAK/STAT3 signaling pathway, which ultimately enhanced the transcription of PD-L1, thus establishing a PD-L1-nPD-L1-PD-L1 feedback loop that ultimately promoted CRC progression and CCLM. In conclusion, our study reveals a novel role for nPD-L1 in CRC, identifies the PD-L1-nPD-L1-PD-L1 feedback loop in CRC, and provides a therapeutic strategy for CRC patients.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
FGFR2 Promotes Expression of PD-L1 in Colorectal Cancer via the JAK/STAT3 Signaling Pathway.J Immunol. 2019 May 15;202(10):3065-3075. doi: 10.4049/jimmunol.1801199. Epub 2019 Apr 12. J Immunol. 2019. PMID: 30979816
-
Nuclear PD-L1 promotes cell cycle progression of BRAF-mutated colorectal cancer by inhibiting THRAP3.Cancer Lett. 2022 Feb 28;527:127-139. doi: 10.1016/j.canlet.2021.12.017. Epub 2021 Dec 17. Cancer Lett. 2022. PMID: 34923044
-
Nuclear PD-L1 promotes EGR1-mediated angiogenesis and accelerates tumorigenesis.Cell Discov. 2023 Mar 28;9(1):33. doi: 10.1038/s41421-023-00521-7. Cell Discov. 2023. PMID: 36977660 Free PMC article.
-
Exosomal PD-L1 derived from head and neck squamous cell carcinoma promotes immune evasion by activating the positive feedback loop of activated regulatory T cell-M2 macrophage.Oral Oncol. 2023 Oct;145:106532. doi: 10.1016/j.oraloncology.2023.106532. Epub 2023 Jul 25. Oral Oncol. 2023. PMID: 37499326 Review.
-
PD-1/PD-L1-dependent immune response in colorectal cancer.J Cell Physiol. 2020 Jul;235(7-8):5461-5475. doi: 10.1002/jcp.29494. Epub 2020 Jan 21. J Cell Physiol. 2020. PMID: 31960962 Review.
References
-
- Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov. 2022;12:134–53. doi: 10.1158/2159-8290.CD-21-0316. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

