Understanding the relationship between cerebellum and the frontal-cortex region of C9orf72-related amyotrophic lateral sclerosis: A comparative analysis of genetic features
- PMID: 38753768
- PMCID: PMC11098475
- DOI: 10.1371/journal.pone.0301267
Understanding the relationship between cerebellum and the frontal-cortex region of C9orf72-related amyotrophic lateral sclerosis: A comparative analysis of genetic features
Abstract
Background: Amyotrophic lateral sclerosis (ALS) is a relentlessly progressive and fatal neurodegenerative diseases for which at present no cure is available. Despite the extensive research the progress from diagnosis to prognosis in ALS and frontotemporal dementia (FTD) has been slow which represents suboptimal understanding of disease pathophysiological processes. In recent studies, several genes have been associated with the ALS and FTD diseases such as SOD1, TDP43, and TBK1, whereas the hexanucleotide GGGGCC repeat expansion (HRE) in C9orf72 gene is a most frequent cause of ALS and FTD, that has changed the understanding of these diseases.
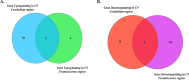
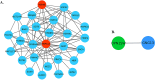
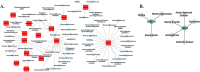
Methods: The goal of this study was to identify and spatially determine differential gene expression signature differences between cerebellum and frontal cortex in C9orf72-associated ALS (C9-ALS), to study the network properties of these differentially expressed genes, and to identify miRNAs targeting the common differentially expressed genes in both the tissues. This study thus highlights underlying differential cell susceptibilities to the disease mechanisms in C9-ALS and suggesting therapeutic target selection in C9-ALS.
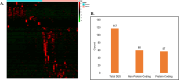
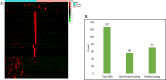
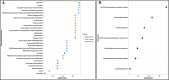
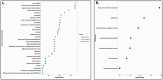
Results: In this manuscript, we have identified that the genes involved in neuron development, protein localization and transcription are mostly enriched in cerebellum of C9-ALS patients, while the UPR-related genes are enriched in the frontal cortex. Of note, UPR pathway genes were mostly dysregulated both in the C9-ALS cerebellum and frontal cortex. Overall, the data presented here show that defects in normal RNA processing and the UPR pathway are the pathological hallmarks of C9-ALS. Interestingly, the cerebellum showed more strong transcriptome changes than the frontal cortex.
Conclusion: Interestingly, the cerebellum region showed more significant transcriptomic changes as compared to the frontal cortex region suggesting its active participation in the disease process. This nuanced understanding may offer valuable insights for the development of targeted therapeutic strategies aimed at mitigating disease progression in C9-ALS.
Copyright: © 2024 Prasad et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS.Acta Neuropathol. 2013 Dec;126(6):829-44. doi: 10.1007/s00401-013-1192-8. Epub 2013 Oct 16. Acta Neuropathol. 2013. PMID: 24129584 Free PMC article.
-
Longitudinal imaging in C9orf72 mutation carriers: Relationship to phenotype.Neuroimage Clin. 2016 Oct 22;12:1035-1043. doi: 10.1016/j.nicl.2016.10.014. eCollection 2016. Neuroimage Clin. 2016. PMID: 27995069 Free PMC article. Clinical Trial.
-
Stable transgenic C9orf72 zebrafish model key aspects of the ALS/FTD phenotype and reveal novel pathological features.Acta Neuropathol Commun. 2018 Nov 19;6(1):125. doi: 10.1186/s40478-018-0629-7. Acta Neuropathol Commun. 2018. PMID: 30454072 Free PMC article.
-
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.Autophagy. 2021 Nov;17(11):3306-3322. doi: 10.1080/15548627.2021.1872189. Epub 2021 Feb 26. Autophagy. 2021. PMID: 33632058 Free PMC article. Review.
-
Synaptic dysfunction and altered excitability in C9ORF72 ALS/FTD.Brain Res. 2018 Aug 15;1693(Pt A):98-108. doi: 10.1016/j.brainres.2018.02.011. Epub 2018 Feb 14. Brain Res. 2018. PMID: 29453960 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

