Metagenomic analysis of sewage for surveillance of bacterial pathogens: A release experiment to determine sensitivity
- PMID: 38753691
- PMCID: PMC11098379
- DOI: 10.1371/journal.pone.0300733
Metagenomic analysis of sewage for surveillance of bacterial pathogens: A release experiment to determine sensitivity
Abstract
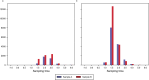
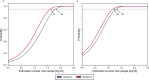
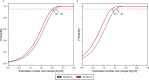
Accurate monitoring of gastro-enteric and other diseases in large populations poses a challenge for public health management. Sewage represents a larger population, is freely obtainable and non-subject to ethical approval. Metagenomic sequencing offers simultaneous, multiple-target analysis. However, no study has demonstrated the sensitivity of metagenomics for detecting bacteria in sewage. In this study, we spot-released 1013 colony-forming units (CFU) of Staphyloccus hyicus (non-pathogenetic strain 842J-88). The strain was flushed down a toilet into the sewer in the catchment area of a public wastewater treatment plant (WWTP), serving a population of 36,000 people. Raw sewage was continuously sampled at the WWTP's inlet over 30- and 60-minute intervals for a total period of seven hours. The experiment was conducted twice with one week in-between release days and under comparable weather conditions. For the metagenomics analyses, the pure single isolate of S. hyicus was sequenced, assembled and added to a large database of bacterial reference sequences. All sewage samples were analyzed by shotgun metagenome sequencing and mapped against the reference database. S. hyicus was identified in duplicate samples at both of two release days and these sequence fragment counts served as a proxy to estimate the minimum number of sick people or sensitivity required in order to observe at least one sick person at 95% probability. We found the sensitivity to be in the range 41-140 and 16-36 sick people at release days 1 and 2, respectively. The WWTP normally serves 36,000 people giving a normalized sensitivity in the range of one in 257 to 2,250 persons.
Copyright: © 2024 Kohle et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Shotgun Metagenomic Profiles Have a High Capacity To Discriminate Samples of Activated Sludge According to Wastewater Type.Appl Environ Microbiol. 2016 Aug 15;82(17):5186-96. doi: 10.1128/AEM.00916-16. Print 2016 Sep 1. Appl Environ Microbiol. 2016. PMID: 27316957 Free PMC article.
-
Time-series sewage metagenomics distinguishes seasonal, human-derived and environmental microbial communities potentially allowing source-attributed surveillance.Nat Commun. 2024 Aug 30;15(1):7551. doi: 10.1038/s41467-024-51957-8. Nat Commun. 2024. PMID: 39215001 Free PMC article.
-
A Metagenomic Investigation of Spatial and Temporal Changes in Sewage Microbiomes across a University Campus.mSystems. 2022 Oct 26;7(5):e0065122. doi: 10.1128/msystems.00651-22. Epub 2022 Sep 19. mSystems. 2022. PMID: 36121163 Free PMC article.
-
Urban metagenomics uncover antibiotic resistance reservoirs in coastal beach and sewage waters.Microbiome. 2019 Feb 28;7(1):35. doi: 10.1186/s40168-019-0648-z. Microbiome. 2019. PMID: 30819245 Free PMC article.
-
Next-generation Sequencing for Surveillance of Antimicrobial Resistance and Pathogenicity in Municipal Wastewater Treatment Plants.Curr Med Chem. 2022;30(1):5-29. doi: 10.2174/0929867329666220802093415. Curr Med Chem. 2022. PMID: 35927898 Review.
References
-
- United States of America Government. Department of Health and Human Services. Centers for Disease Control and Prevention (2015). FoodNet Surveillance. Burden of Illness. Retrieved from https://tinyurl.com/ufj9vu8 on the 21 January 2020.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

