This is a preprint.
Cysteine Rich Intestinal Protein 2 is a copper-responsive regulator of skeletal muscle differentiation
- PMID: 38746126
- PMCID: PMC11092763
- DOI: 10.1101/2024.05.03.592485
Cysteine Rich Intestinal Protein 2 is a copper-responsive regulator of skeletal muscle differentiation
Abstract
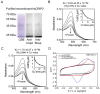
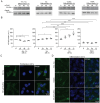
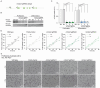
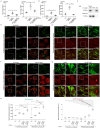
Copper (Cu) is an essential trace element required for respiration, neurotransmitter synthesis, oxidative stress response, and transcriptional regulation. Imbalance in Cu homeostasis can lead to several pathological conditions, affecting neuronal, cognitive, and muscular development. Mechanistically, Cu and Cu-binding proteins (Cu-BPs) have an important but underappreciated role in transcription regulation in mammalian cells. In this context, our lab investigates the contributions of novel Cu-BPs in skeletal muscle differentiation using murine primary myoblasts. Through an unbiased synchrotron X-ray fluorescence-mass spectrometry (XRF/MS) metalloproteomic approach, we identified the murine cysteine rich intestinal protein 2 (mCrip2) in a sample that showed enriched Cu signal, which was isolated from differentiating primary myoblasts derived from mouse satellite cells. Immunolocalization analyses showed that mCrip2 is abundant in both nuclear and cytosolic fractions. Thus, we hypothesized that mCrip2 might have differential roles depending on its cellular localization in the skeletal muscle lineage. mCrip2 is a LIM-family protein with 4 conserved Zn2+-binding sites. Homology and phylogenetic analyses showed that mammalian Crip2 possesses histidine residues near two of the Zn2+-binding sites (CX2C-HX2C) which are potentially implicated in Cu+-binding and competition with Zn2+. Biochemical characterization of recombinant human hsCRIP2 revealed a high Cu+-binding affinity for two and four Cu+ ions and limited redox potential. Functional characterization using CRISPR/Cas9-mediated deletion of mCrip2 in primary myoblasts did not impact proliferation, but impaired myogenesis by decreasing the expression of differentiation markers, possibly attributed to Cu accumulation. Transcriptome analyses of proliferating and differentiating mCrip2 KO myoblasts showed alterations in mRNA processing, protein translation, ribosome synthesis, and chromatin organization. CUT&RUN analyses showed that mCrip2 associates with a select set of gene promoters, including MyoD1 and metallothioneins, acting as a novel Cu-responsive or Cu-regulating protein. Our work demonstrates novel regulatory functions of mCrip2 that mediate skeletal muscle differentiation, presenting new features of the Cu-network in myoblasts.
Keywords: Crip2; copper; myogenesis; transcriptional regulation.
Figures



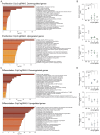
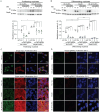
Similar articles
-
The classic metal-sensing transcription factor MTF1 promotes myogenesis in response to copper.FASEB J. 2019 Dec;33(12):14556-14574. doi: 10.1096/fj.201901606R. Epub 2019 Nov 5. FASEB J. 2019. PMID: 31690123 Free PMC article.
-
BosR Is A Novel Fur Family Member Responsive to Copper and Regulating Copper Homeostasis in Borrelia burgdorferi.J Bacteriol. 2017 Jul 25;199(16):e00276-17. doi: 10.1128/JB.00276-17. Print 2017 Aug 15. J Bacteriol. 2017. PMID: 28583949 Free PMC article.
-
Dynamic changes in copper homeostasis and post-transcriptional regulation of Atp7a during myogenic differentiation.Metallomics. 2018 Feb 21;10(2):309-322. doi: 10.1039/c7mt00324b. Metallomics. 2018. PMID: 29333545 Free PMC article.
-
Growth hormone and the insulin-like growth factor system in myogenesis.Endocr Rev. 1996 Oct;17(5):481-517. doi: 10.1210/edrv-17-5-481. Endocr Rev. 1996. PMID: 8897022 Review.
-
RNA-Binding Proteins in the Post-transcriptional Control of Skeletal Muscle Development, Regeneration and Disease.Front Cell Dev Biol. 2021 Sep 20;9:738978. doi: 10.3389/fcell.2021.738978. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34616743 Free PMC article. Review.
References
-
- Fraústo da Silva J. J. R., and Williams R. J. P. (2001) The biological chemistry of the elements : the inorganic chemistry of life, Oxford University Press, Oxford
-
- Csiszar K. (2001) Lysyl oxidases: a novel multifunctional amine oxidase family. Progress in nucleic acid research and molecular biology 70, 1–32 - PubMed
-
- Moriya M., Ho Y. H., Grana A., Nguyen L., Alvarez A., Jamil R., Ackland M. L., Michalczyk A., Hamer P., Ramos D., Kim S., Mercer J. F., and Linder M. C. (2008) Copper is taken up efficiently from albumin and alpha2-macroglobulin by cultured human cells by more than one mechanism. American journal of physiology. Cell physiology 295, C708–721 - PMC - PubMed
-
- Chen L., Li N., Zhang M., Sun M., Bian J., Yang B., Li Z., Wang J., Li F., Shi X., Wang Y., Yuan F., Zou P., Shan C., and Wang J. (2021) APEX2-based Proximity Labeling of Atox1 Identifies CRIP2 as a Nuclear Copper-binding Protein that Regulates Autophagy Activation. Angew Chem Int Ed Engl 60, 25346–25355 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
