SARS-CoV-2 aberrantly elevates mitochondrial bioenergetics to induce robust virus propagation
- PMID: 38734691
- PMCID: PMC11088672
- DOI: 10.1038/s41392-024-01836-x
SARS-CoV-2 aberrantly elevates mitochondrial bioenergetics to induce robust virus propagation
Abstract
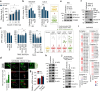
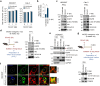
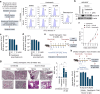
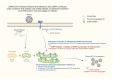
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a 'highly transmissible respiratory pathogen, leading to severe multi-organ damage. However, knowledge regarding SARS-CoV-2-induced cellular alterations is limited. In this study, we report that SARS-CoV-2 aberrantly elevates mitochondrial bioenergetics and activates the EGFR-mediated cell survival signal cascade during the early stage of viral infection. SARS-CoV-2 causes an increase in mitochondrial transmembrane potential via the SARS-CoV-2 RNA-nucleocapsid cluster, thereby abnormally promoting mitochondrial elongation and the OXPHOS process, followed by enhancing ATP production. Furthermore, SARS-CoV-2 activates the EGFR signal cascade and subsequently induces mitochondrial EGFR trafficking, contributing to abnormal OXPHOS process and viral propagation. Approved EGFR inhibitors remarkably reduce SARS-CoV-2 propagation, among which vandetanib exhibits the highest antiviral efficacy. Treatment of SARS-CoV-2-infected cells with vandetanib decreases SARS-CoV-2-induced EGFR trafficking to the mitochondria and restores SARS-CoV-2-induced aberrant elevation in OXPHOS process and ATP generation, thereby resulting in the reduction of SARS-CoV-2 propagation. Furthermore, oral administration of vandetanib to SARS-CoV-2-infected hACE2 transgenic mice reduces SARS-CoV-2 propagation in lung tissue and mitigates SARS-CoV-2-induced lung inflammation. Vandetanib also exhibits potent antiviral activity against various SARS-CoV-2 variants of concern, including alpha, beta, delta and omicron, in in vitro cell culture experiments. Taken together, our findings provide novel insight into SARS-CoV-2-induced alterations in mitochondrial dynamics and EGFR trafficking during the early stage of viral infection and their roles in robust SARS-CoV-2 propagation, suggesting that EGFR is an attractive host target for combating COVID-19.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Reduction of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant infection by blocking the epidermal growth factor receptor (EGFR) pathway.Microbiol Spectr. 2024 Nov 5;12(11):e0158324. doi: 10.1128/spectrum.01583-24. Epub 2024 Sep 18. Microbiol Spectr. 2024. PMID: 39291996 Free PMC article.
-
Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice.Virol J. 2021 Feb 27;18(1):46. doi: 10.1186/s12985-021-01515-1. Virol J. 2021. PMID: 33639976 Free PMC article.
-
Effects of simeprevir on the replication of SARS-CoV-2 in vitro and in transgenic hACE2 mice.Int J Antimicrob Agents. 2022 Jan;59(1):106499. doi: 10.1016/j.ijantimicag.2021.106499. Epub 2021 Dec 17. Int J Antimicrob Agents. 2022. PMID: 34929295 Free PMC article.
-
SARS-CoV-2 mitochondrial metabolic and epigenomic reprogramming in COVID-19.Pharmacol Res. 2024 Jun;204:107170. doi: 10.1016/j.phrs.2024.107170. Epub 2024 Apr 12. Pharmacol Res. 2024. PMID: 38614374 Review.
-
Severe acute respiratory syndrome coronaviruses contributing to mitochondrial dysfunction: Implications for post-COVID complications.Mitochondrion. 2023 Mar;69:43-56. doi: 10.1016/j.mito.2023.01.005. Epub 2023 Jan 20. Mitochondrion. 2023. PMID: 36690315 Free PMC article. Review.
Cited by
-
Aging mitochondria in the context of SARS-CoV-2: exploring interactions and implications.Front Aging. 2024 Sep 24;5:1442323. doi: 10.3389/fragi.2024.1442323. eCollection 2024. Front Aging. 2024. PMID: 39380657 Free PMC article. Review.
-
Post-Acute Sequelae and Mitochondrial Aberration in SARS-CoV-2 Infection.Int J Mol Sci. 2024 Aug 21;25(16):9050. doi: 10.3390/ijms25169050. Int J Mol Sci. 2024. PMID: 39201736 Free PMC article. Review.
-
Viruses and Mitochondrial Dysfunction in Neurodegeneration and Cognition: An Evolutionary Perspective.Cell Mol Neurobiol. 2024 Oct 17;44(1):68. doi: 10.1007/s10571-024-01503-3. Cell Mol Neurobiol. 2024. PMID: 39417916 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- CRC-16-01-KRICT/National Research Council of Science and Technology (National Research Council of Science & Technology)
- KK2333-20/Korea Research Institute of Chemical Technology (KRICT)
- NRF-2021M3E5E3080540/National Research Foundation of Korea (NRF)
- RS-2023-00248135/National Research Foundation of Korea (NRF)
- BK 21 FOUR Program by the Chungnam National University Research Grant, 2023/Chungnam National University (CNU)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

