Regulation of Myc transcription by an enhancer cluster dedicated to pluripotency and early embryonic expression
- PMID: 38729993
- PMCID: PMC11087473
- DOI: 10.1038/s41467-024-48258-5
Regulation of Myc transcription by an enhancer cluster dedicated to pluripotency and early embryonic expression
Abstract
MYC plays various roles in pluripotent stem cells, including the promotion of somatic cell reprogramming to pluripotency, the regulation of cell competition and the control of embryonic diapause. However, how Myc expression is regulated in this context remains unknown. The Myc gene lies within a ~ 3-megabase gene desert with multiple cis-regulatory elements. Here we use genomic rearrangements, transgenesis and targeted mutation to analyse Myc regulation in early mouse embryos and pluripotent stem cells. We identify a topologically-associated region that homes enhancers dedicated to Myc transcriptional regulation in stem cells of the pre-implantation and early post-implantation embryo. Within this region, we identify elements exclusively dedicated to Myc regulation in pluripotent cells, with distinct enhancers that sequentially activate during naive and formative pluripotency. Deletion of pluripotency-specific enhancers dampens embryonic stem cell competitive ability. These results identify a topologically defined enhancer cluster dedicated to early embryonic expression and uncover a modular mechanism for the regulation of Myc expression in different states of pluripotency.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


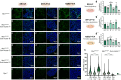



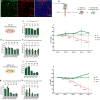
Similar articles
-
Molecular coupling of Tsix regulation and pluripotency.Nature. 2010 Nov 18;468(7322):457-60. doi: 10.1038/nature09496. Nature. 2010. PMID: 21085182
-
Overexpression Nanog activates pluripotent genes in porcine fetal fibroblasts and nuclear transfer embryos.Anat Rec (Hoboken). 2011 Nov;294(11):1809-17. doi: 10.1002/ar.21457. Epub 2011 Oct 3. Anat Rec (Hoboken). 2011. PMID: 21972213
-
Pluripotency Surveillance by Myc-Driven Competitive Elimination of Differentiating Cells.Dev Cell. 2017 Sep 25;42(6):585-599.e4. doi: 10.1016/j.devcel.2017.08.011. Epub 2017 Sep 14. Dev Cell. 2017. PMID: 28919206
-
Transcriptional Control of Somatic Cell Reprogramming.Trends Cell Biol. 2016 Apr;26(4):272-288. doi: 10.1016/j.tcb.2015.12.003. Epub 2016 Jan 15. Trends Cell Biol. 2016. PMID: 26776886 Review.
-
Mechanisms of gene regulation in human embryos and pluripotent stem cells.Development. 2017 Dec 15;144(24):4496-4509. doi: 10.1242/dev.157404. Development. 2017. PMID: 29254992 Free PMC article. Review.
Cited by
-
The evolution of multicellularity and cell differentiation symposium: bridging evolutionary cell biology and computational modelling using emerging model systems.Biol Open. 2024 Jul 15;13(10):bio061720. doi: 10.1242/bio.061720. Epub 2024 Oct 7. Biol Open. 2024. PMID: 39373528 Free PMC article.
-
Skin Development and Disease: A Molecular Perspective.Curr Issues Mol Biol. 2024 Jul 30;46(8):8239-8267. doi: 10.3390/cimb46080487. Curr Issues Mol Biol. 2024. PMID: 39194704 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- PGC2018-096486-B-I00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- CEX2020-001041-S/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- SC1-BHC-07-2019/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- P2022/BMD-7245 CARDIOBOOST-CM/Comunidad de Madrid
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

