UV damage induces production of mitochondrial DNA fragments with specific length profiles
- PMID: 38722894
- PMCID: PMC11228841
- DOI: 10.1093/genetics/iyae070
UV damage induces production of mitochondrial DNA fragments with specific length profiles
Abstract
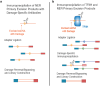
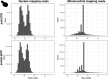
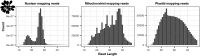
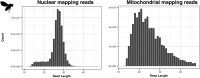
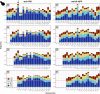
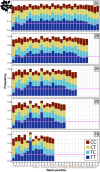
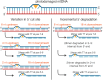
UV light is a potent mutagen that induces bulky DNA damage in the form of cyclobutane pyrimidine dimers (CPDs). Photodamage and other bulky lesions occurring in nuclear genomes can be repaired through nucleotide excision repair (NER), where incisions on both sides of a damaged site precede the removal of a single-stranded oligonucleotide containing the damage. Mitochondrial genomes (mtDNAs) are also susceptible to damage from UV light, but current evidence suggests that the only way to eliminate bulky mtDNA damage is through mtDNA degradation. Damage-containing oligonucleotides excised during NER can be captured with antidamage antibodies and sequenced (XR-seq) to produce high-resolution maps of active repair locations following UV exposure. We analyzed previously published datasets from Arabidopsis thaliana, Saccharomyces cerevisiae, and Drosophila melanogaster to identify reads originating from the mtDNA (and plastid genome in A. thaliana). In A. thaliana and S. cerevisiae, the mtDNA-mapping reads have unique length distributions compared to the nuclear-mapping reads. The dominant fragment size was 26 nt in S. cerevisiae and 28 nt in A. thaliana with distinct secondary peaks occurring in regular intervals. These reads also show a nonrandom distribution of di-pyrimidines (the substrate for CPD formation) with TT enrichment at positions 7-8 of the reads. Therefore, UV damage to mtDNA appears to result in production of DNA fragments of characteristic lengths and positions relative to the damaged location. The mechanisms producing these fragments are unclear, but we hypothesize that they result from a previously uncharacterized DNA degradation pathway or repair mechanism in mitochondria.
Keywords: Arabidopsis thaliana; Saccharomyces cerevisiae; XR sequencing; cyclobutane pyrimidine dimer (CPD); mitochondrial genome (mtDNA); mtDNA degradation; nucleotide excision repair (NER); photodamage.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflicts of interest.
Figures
Update of
-
UV damage induces production of mitochondrial DNA fragments with specific length profiles.bioRxiv [Preprint]. 2023 Nov 11:2023.11.07.566130. doi: 10.1101/2023.11.07.566130. bioRxiv. 2023. Update in: Genetics. 2024 Jul 8;227(3):iyae070. doi: 10.1093/genetics/iyae070. PMID: 37986892 Free PMC article. Updated. Preprint.
Similar articles
-
UV damage induces production of mitochondrial DNA fragments with specific length profiles.bioRxiv [Preprint]. 2023 Nov 11:2023.11.07.566130. doi: 10.1101/2023.11.07.566130. bioRxiv. 2023. Update in: Genetics. 2024 Jul 8;227(3):iyae070. doi: 10.1093/genetics/iyae070. PMID: 37986892 Free PMC article. Updated. Preprint.
-
Erratum: Eyestalk Ablation to Increase Ovarian Maturation in Mud Crabs.J Vis Exp. 2023 May 26;(195). doi: 10.3791/6561. J Vis Exp. 2023. PMID: 37235796
-
Discarded sequencing reads uncover natural variation in pest resistance in Thlaspi arvense.Elife. 2024 Dec 19;13:RP95510. doi: 10.7554/eLife.95510. Elife. 2024. PMID: 39699583 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article. Review.
References
-
- Akkose U, Kaya VO, Lindsey-Boltz L, Karagoz Z, Brown AD, Larsen PA, Yoder AD, Sancar A, Adebali O. 2021. Comparative analyses of two primate species diverged by more than 60 million years show different rates but similar distribution of genome-wide UV repair events. BMC Genomics. 22(1):600. doi:10.1186/s12864-021-07898-3. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

