An ALS-associated mutation dysregulates microglia-derived extracellular microRNAs in a sex-specific manner
- PMID: 38721655
- PMCID: PMC11152562
- DOI: 10.1242/dmm.050638
An ALS-associated mutation dysregulates microglia-derived extracellular microRNAs in a sex-specific manner
Abstract
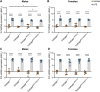
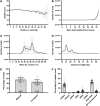
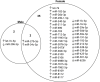
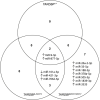
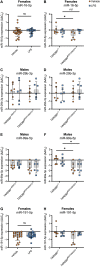
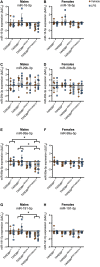
Evidence suggests the presence of microglial activation and microRNA (miRNA) dysregulation in amyotrophic lateral sclerosis (ALS), the most common form of adult motor neuron disease. However, few studies have investigated whether the miRNA dysregulation originates from microglia. Furthermore, TDP-43 (encoded by TARDBP), involved in miRNA biogenesis, aggregates in tissues of ∼98% of ALS cases. Thus, this study aimed to determine whether expression of the ALS-linked TDP-43M337V mutation in a transgenic mouse model dysregulates microglia-derived miRNAs. RNA sequencing identified several dysregulated miRNAs released by transgenic microglia and a differential miRNA release by lipopolysaccharide-stimulated microglia, which was more pronounced in cells from female mice. We validated the downregulation of three candidate miRNAs, namely, miR-16-5p, miR-99a-5p and miR-191-5p, by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and identified their predicted targets, which primarily include genes involved in neuronal development and function. These results suggest that altered TDP-43 function leads to changes in the miRNA population released by microglia, which may in turn be a source of the miRNA dysregulation observed in the disease. This has important implications for the role of neuroinflammation in ALS pathology and could provide potential therapeutic targets.
Keywords: Amyotrophic lateral sclerosis; MicroRNA; Microglia; Motor neuron disease; TDP-43.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests E.C. holds shares in Thermo Fisher Scientific and various index funds, which may include companies whose products were used in the research reported in this article. Additionally, E.C. has been engaged in a compensated collaboration with Thermo Fisher Scientific for promotional activities unrelated to this research. These potential financial interests do not influence the design, execution or interpretation of the research findings presented in this article. All other authors declare no competing interests.
Figures

Similar articles
-
Systemic dysregulation of TDP-43 binding microRNAs in amyotrophic lateral sclerosis.Acta Neuropathol Commun. 2013 Jul 30;1:42. doi: 10.1186/2051-5960-1-42. Acta Neuropathol Commun. 2013. PMID: 24252274 Free PMC article.
-
Potential of activated microglia as a source of dysregulated extracellular microRNAs contributing to neurodegeneration in amyotrophic lateral sclerosis.J Neuroinflammation. 2020 Apr 28;17(1):135. doi: 10.1186/s12974-020-01822-4. J Neuroinflammation. 2020. PMID: 32345319 Free PMC article. Review.
-
A Panel of miRNA Biomarkers Common to Serum and Brain-Derived Extracellular Vesicles Identified in Mouse Model of Amyotrophic Lateral Sclerosis.Mol Neurobiol. 2024 Aug;61(8):5901-5915. doi: 10.1007/s12035-023-03857-z. Epub 2024 Jan 22. Mol Neurobiol. 2024. PMID: 38252383 Free PMC article.
-
Novel miR-b2122 regulates several ALS-related RNA-binding proteins.Mol Brain. 2017 Oct 2;10(1):46. doi: 10.1186/s13041-017-0326-7. Mol Brain. 2017. PMID: 28969660 Free PMC article.
-
MicroRNA Metabolism and Dysregulation in Amyotrophic Lateral Sclerosis.Mol Neurobiol. 2018 Mar;55(3):2617-2630. doi: 10.1007/s12035-017-0537-z. Epub 2017 Apr 18. Mol Neurobiol. 2018. PMID: 28421535 Review.
References
-
- Afgan, E., Baker, D., Batut, B., van den beek, M., Bouvier, D., Čech, M., Chilton, J., Clements, D., Coraor, N., Grüning, B. A.et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537-W544. 10.1093/nar/gky379 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

