This is a preprint.
A first comprehensive analysis of Transcribed Ultra Conserved Regions uncovers important regulatory functions of novel non-coding transcripts in gliomas
- PMID: 38699302
- PMCID: PMC11065071
- DOI: 10.21203/rs.3.rs-4164642/v1
A first comprehensive analysis of Transcribed Ultra Conserved Regions uncovers important regulatory functions of novel non-coding transcripts in gliomas
Abstract
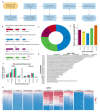
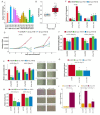
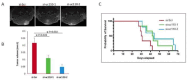
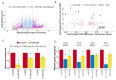
Transcribed Ultra-Conserved Regions (TUCRs) represent a severely understudied class of putative non-coding RNAs (ncRNAs) that are 100% conserved across multiple species. We performed the first-ever analysis of TUCRs in glioblastoma (GBM) and low-grade gliomas (LGG). We leveraged large human datasets to identify the genomic locations, chromatin accessibility, transcription, differential expression, correlation with survival, and predicted functions of all 481 TUCRs, and identified TUCRs that are relevant to glioma biology. Of these, we investigated the expression, function, and mechanism of action of the most highly upregulated intergenic TUCR, uc.110, identifying it as a new oncogene. Uc.110 was highly overexpressed in GBM and LGG, where it promoted malignancy and tumor growth. Uc.110 activated the WNT pathway by upregulating the expression of membrane frizzled-related protein (MFRP), by sponging the tumor suppressor microRNA miR-544. This pioneering study shows important roles for TUCRs in gliomas and provides an extensive database and novel methods for future TUCR research.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures


Similar articles
-
A comprehensive analysis of Transcribed Ultra Conserved Regions uncovers important regulatory functions of novel non-coding transcripts in gliomas.bioRxiv [Preprint]. 2024 Dec 22:2023.09.12.557444. doi: 10.1101/2023.09.12.557444. bioRxiv. 2024. PMID: 38562826 Free PMC article. Preprint.
-
TUCR.454 inhabits metastasis in lung cancer cells.Int J Clin Exp Pathol. 2018 Mar 1;11(3):1289-1296. eCollection 2018. Int J Clin Exp Pathol. 2018. PMID: 31938224 Free PMC article.
-
Transcribed Ultraconserved Regions in Cancer.Cells. 2022 May 19;11(10):1684. doi: 10.3390/cells11101684. Cells. 2022. PMID: 35626721 Free PMC article. Review.
-
DKK3 expression is associated with immunosuppression and poor prognosis in glioblastoma, in contrast to lower-grade gliomas.BMC Neurol. 2023 May 6;23(1):183. doi: 10.1186/s12883-023-03236-0. BMC Neurol. 2023. PMID: 37149563 Free PMC article.
-
Non-coding RNAs and glioblastoma: Insight into their roles in metastasis.Mol Ther Oncolytics. 2021 Dec 22;24:262-287. doi: 10.1016/j.omto.2021.12.015. eCollection 2022 Mar 17. Mol Ther Oncolytics. 2021. PMID: 35071748 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

