Isolation of quinic acid from dropped Citrus reticulata Blanco fruits: its derivatization, antibacterial potential, docking studies, and ADMET profiling
- PMID: 38698937
- PMCID: PMC11064019
- DOI: 10.3389/fchem.2024.1372560
Isolation of quinic acid from dropped Citrus reticulata Blanco fruits: its derivatization, antibacterial potential, docking studies, and ADMET profiling
Abstract
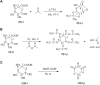
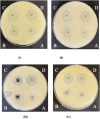
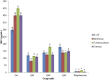
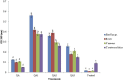
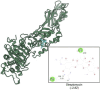
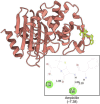
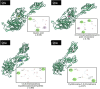
Citrus reticulata dropped fruits are generally discarded as waste, causing environmental pollution and losses to farmers. In the present study, column chromatography has been used to isolate quinic acid (1,3,4,5-tetrahydroxycyclohexane-1-carboxylic acid) from the ethyl acetate fraction of a methanol extract of citrus fruits dropped in April. Quinic acid is a ubiquitous plant metabolite found in various plants and microorganisms. It is an important precursor in the biosynthesis of aromatic natural compounds. It was further derivatized into 3,4-o-isopropylidenequinic acid 1,5-lactone (QA1), 1,3,4,5-tetraacetoxycyclohexylaceticanhydride (QA2), and cyclohexane-1,2,3,5-tetraone (QA3). These compounds were further tested for their antibacterial potential against the foodborne pathogens Staphylococcus aureus, Bacillus spp., Yersinia enterocolitica, and Escherichia coli. QA1 exhibited maximum antibacterial potential (minimum inhibitory concentration; 80-120 μg/mL). QA1 revealed synergistic behavior with streptomycin against all the tested bacterial strains having a fractional inhibitory concentration index ranging from 0.29 to 0.37. It also caused a significant increase in cell constituent release in all the tested bacteria compared to the control, along with prominent biofilm reduction. The results obtained were further checked with computational studies that revealed the best docking score of QA1 (-6.30 kcal/mol, -5.8 kcal/mol, and -4.70 kcal/mol) against β-lactamase, DNA gyrase, and transpeptidase, respectively. The absorption, distribution, metabolism, excretion, and toxicity (ADMET) analysis revealed that the drug-like properties of QA1 had an ideal toxicity profile, making it a suitable candidate for the development of antimicrobial drugs.
Keywords: biofilm; citrus reticulata; docking; dropped citrus fruits; lactone; quinic acid.
Copyright © 2024 Heena, Kaushal, Kaur, Panwar, Sharma and Jangra.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
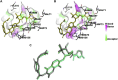


Similar articles
-
Chemical Composition and Biological Activities of the Essential Oil from Citrus reticulata Blanco Peels Collected from Agrowastes.Chem Biodivers. 2024 Mar;21(3):e202301223. doi: 10.1002/cbdv.202301223. Epub 2024 Feb 27. Chem Biodivers. 2024. PMID: 38108562
-
Quantification of Flavonoids, Phenols and Antioxidant Potential from Dropped Citrus reticulata Blanco Fruits Influenced by Drying Techniques.Molecules. 2021 Jul 8;26(14):4159. doi: 10.3390/molecules26144159. Molecules. 2021. PMID: 34299432 Free PMC article.
-
Evaluation of Skin Anti-aging Potential of Citrus reticulata Blanco Peel.Pharmacognosy Res. 2016 Jul-Sep;8(3):160-8. doi: 10.4103/0974-8490.182913. Pharmacognosy Res. 2016. PMID: 27365982 Free PMC article.
-
Antibacterial and antioxidant activities of extracts and isolated compounds from the roots extract of Cucumis prophetarum and in silico study on DNA gyrase and human peroxiredoxin 5.BMC Chem. 2021 May 6;15(1):32. doi: 10.1186/s13065-021-00758-x. BMC Chem. 2021. PMID: 33957962 Free PMC article.
-
The Chemical Compositions, and Antibacterial and Antioxidant Activities of Four Types of Citrus Essential Oils.Molecules. 2021 Jun 4;26(11):3412. doi: 10.3390/molecules26113412. Molecules. 2021. PMID: 34199966 Free PMC article.
Cited by
-
Bioactive Compounds and Valorization of Coffee By-Products from the Origin: A Circular Economy Model from Local Practices in Zongolica, Mexico.Plants (Basel). 2024 Sep 30;13(19):2741. doi: 10.3390/plants13192741. Plants (Basel). 2024. PMID: 39409611 Free PMC article.
References
-
- Arya A., Al-Obaidi M. M. J., Shahid N., Noordin M. I. B., Looi C. Y., Wong W. F., et al. (2014). Synergistic effect of quercetin and quinic acid by alleviating structural degeneration in the liver, kidney and pancreas tissues of STZ-induced diabetic rats: a mechanistic study. Food Chem. Toxicol. 71, 183–196. 10.1016/j.fct.2014.06.010 - DOI - PubMed
-
- Bai J., Wu Y., Wang X., Liu X., Zhong K., Huang Y., et al. (2018). In vitro and in vivo characterization of the antibacterial activity and membrane damage mechanism of quinic acid against Staphylococcus aureus . J. Food Saf. 38 (1), e12416. 10.1111/jfs.12416 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

