From genome to evolution: investigating type II methylotrophs using a pangenomic analysis
- PMID: 38695578
- PMCID: PMC11237726
- DOI: 10.1128/msystems.00248-24
From genome to evolution: investigating type II methylotrophs using a pangenomic analysis
Abstract
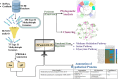
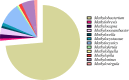
A comprehensive pangenomic approach was employed to analyze the genomes of 75 type II methylotrophs spanning various genera. Our investigation revealed 256 exact core gene families shared by all 75 organisms, emphasizing their crucial role in the survival and adaptability of these organisms. Additionally, we predicted the functionality of 12 hypothetical proteins. The analysis unveiled a diverse array of genes associated with key metabolic pathways, including methane, serine, glyoxylate, and ethylmalonyl-CoA (EMC) metabolic pathways. While all selected organisms possessed essential genes for the serine pathway, Methylooceanibacter marginalis lacked serine hydroxymethyltransferase (SHMT), and Methylobacterium variabile exhibited both isozymes of SHMT, suggesting its potential to utilize a broader range of carbon sources. Notably, Methylobrevis sp. displayed a unique serine-glyoxylate transaminase isozyme not found in other organisms. Only nine organisms featured anaplerotic enzymes (isocitrate lyase and malate synthase) for the glyoxylate pathway, with the rest following the EMC pathway. Methylovirgula sp. 4MZ18 stood out by acquiring genes from both glyoxylate and EMC pathways, and Methylocapsa sp. S129 featured an A-form malate synthase, unlike the G-form found in the remaining organisms. Our findings also revealed distinct phylogenetic relationships and clustering patterns among type II methylotrophs, leading to the proposal of a separate genus for Methylovirgula sp. 4M-Z18 and Methylocapsa sp. S129. This pangenomic study unveils remarkable metabolic diversity, unique gene characteristics, and distinct clustering patterns of type II methylotrophs, providing valuable insights for future carbon sequestration and biotechnological applications.
Importance: Methylotrophs have played a significant role in methane-based product production for many years. However, a comprehensive investigation into the diverse genetic architectures across different genera of methylotrophs has been lacking. This study fills this knowledge gap by enhancing our understanding of core hypothetical proteins and unique enzymes involved in methane oxidation, serine, glyoxylate, and ethylmalonyl-CoA pathways. These findings provide a valuable reference for researchers working with other methylotrophic species. Furthermore, this study not only unveils distinctive gene characteristics and phylogenetic relationships but also suggests a reclassification for Methylovirgula sp. 4M-Z18 and Methylocapsa sp. S129 into separate genera due to their unique attributes within their respective genus. Leveraging the synergies among various methylotrophic organisms, the scientific community can potentially optimize metabolite production, increasing the yield of desired end products and overall productivity.
Keywords: PPanGOLLiN; hypothetical proteins; isozymes; methane; persistent; serine pathway.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The apparent malate synthase activity of Rhodobacter sphaeroides is due to two paralogous enzymes, (3S)-Malyl-coenzyme A (CoA)/{beta}-methylmalyl-CoA lyase and (3S)- Malyl-CoA thioesterase.J Bacteriol. 2010 Mar;192(5):1249-58. doi: 10.1128/JB.01267-09. Epub 2010 Jan 4. J Bacteriol. 2010. PMID: 20047909 Free PMC article.
-
Ethylmalonyl coenzyme A mutase operates as a metabolic control point in Methylobacterium extorquens AM1.J Bacteriol. 2015 Feb 15;197(4):727-35. doi: 10.1128/JB.02478-14. Epub 2014 Dec 1. J Bacteriol. 2015. PMID: 25448820 Free PMC article.
-
Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics.Proc Natl Acad Sci U S A. 2009 Mar 24;106(12):4846-51. doi: 10.1073/pnas.0810932106. Epub 2009 Mar 4. Proc Natl Acad Sci U S A. 2009. PMID: 19261854 Free PMC article.
-
How half a century of research was required to understand bacterial growth on C1 and C2 compounds; the story of the serine cycle and the ethylmalonyl-CoA pathway.Sci Prog. 2011;94(Pt 2):109-37. doi: 10.3184/003685011X13044430633960. Sci Prog. 2011. PMID: 21805909 Free PMC article. Review.
-
Biotechnological potential of the ethylmalonyl-CoA pathway.Appl Microbiol Biotechnol. 2011 Jan;89(1):17-25. doi: 10.1007/s00253-010-2873-z. Epub 2010 Sep 30. Appl Microbiol Biotechnol. 2011. PMID: 20882276 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

