Modeling spillover dynamics: understanding emerging pathogens of public health concern
- PMID: 38684927
- PMCID: PMC11058258
- DOI: 10.1038/s41598-024-60661-y
Modeling spillover dynamics: understanding emerging pathogens of public health concern
Abstract
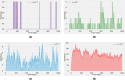
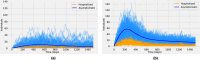
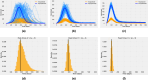
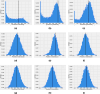
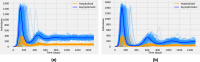
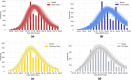
The emergence of infectious diseases with pandemic potential is a major public health threat worldwide. The World Health Organization reports that about 60% of emerging infectious diseases are zoonoses, originating from spillover events. Although the mechanisms behind spillover events remain unclear, mathematical modeling offers a way to understand the intricate interactions among pathogens, wildlife, humans, and their shared environment. Aiming at gaining insights into the dynamics of spillover events and the outcome of an eventual disease outbreak in a population, we propose a continuous time stochastic modeling framework. This framework links the dynamics of animal reservoirs and human hosts to simulate cross-species disease transmission. We conduct a thorough analysis of the model followed by numerical experiments that explore various spillover scenarios. The results suggest that although most epidemic outbreaks caused by novel zoonotic pathogens do not persist in the human population, the rising number of spillover events can avoid long-lasting extinction and lead to unexpected large outbreaks. Hence, global efforts to reduce the impacts of emerging diseases should not only address post-emergence outbreak control but also need to prevent pandemics before they are established.
Keywords: Disease modeling; Emerging infectious diseases; Spillover.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The Vietnam Initiative on Zoonotic Infections (VIZIONS): A Strategic Approach to Studying Emerging Zoonotic Infectious Diseases.Ecohealth. 2015 Dec;12(4):726-35. doi: 10.1007/s10393-015-1061-0. Epub 2015 Sep 24. Ecohealth. 2015. PMID: 26403795 Free PMC article. Review.
-
Stochastic dynamics of an epidemic with recurrent spillovers from an endemic reservoir.J Theor Biol. 2018 Nov 14;457:37-50. doi: 10.1016/j.jtbi.2018.08.017. Epub 2018 Aug 16. J Theor Biol. 2018. PMID: 30121292 Free PMC article.
-
Nine challenges in modelling the emergence of novel pathogens.Epidemics. 2015 Mar;10:35-9. doi: 10.1016/j.epidem.2014.09.002. Epub 2014 Sep 19. Epidemics. 2015. PMID: 25843380 Free PMC article.
-
Spillover and pandemic properties of zoonotic viruses with high host plasticity.Sci Rep. 2015 Oct 7;5:14830. doi: 10.1038/srep14830. Sci Rep. 2015. PMID: 26445169 Free PMC article.
-
Synthesizing the connections between environmental disturbances and zoonotic spillover.An Acad Bras Cienc. 2022 Sep 19;94(suppl 3):e20211530. doi: 10.1590/0001-3765202220211530. eCollection 2022. An Acad Bras Cienc. 2022. PMID: 36169531 Review.
Cited by
-
Modelling COVID-19 mutant dynamics: understanding the interplay between viral evolution and disease transmission dynamics.R Soc Open Sci. 2024 Oct 30;11(10):240919. doi: 10.1098/rsos.240919. eCollection 2024 Oct. R Soc Open Sci. 2024. PMID: 39493297 Free PMC article.
References
-
- Evans T, et al. Links between ecological integrity, emerging infectious diseases originating from wildlife, and other aspects of human health-an overview of the literature. Wildl. Conserv. Soc. 2020;4:e303. doi: 10.13140/RG.2.2.34736.51205. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

