Transcriptional Responses of Different Brain Cell Types to Oxygen Decline
- PMID: 38671993
- PMCID: PMC11048388
- DOI: 10.3390/brainsci14040341
Transcriptional Responses of Different Brain Cell Types to Oxygen Decline
Abstract
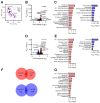
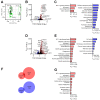
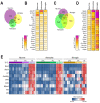
Brain hypoxia is associated with a wide range of physiological and clinical conditions. Although oxygen is an essential constituent of maintaining brain functions, our understanding of how specific brain cell types globally respond and adapt to decreasing oxygen conditions is incomplete. In this study, we exposed mouse primary neurons, astrocytes, and microglia to normoxia and two hypoxic conditions and obtained genome-wide transcriptional profiles of the treated cells. Analysis of differentially expressed genes under conditions of reduced oxygen revealed a canonical hypoxic response shared among different brain cell types. In addition, we observed a higher sensitivity of neurons to oxygen decline, and dissected cell type-specific biological processes affected by hypoxia. Importantly, this study establishes novel gene modules associated with brain cells responding to oxygen deprivation and reveals a state of profound stress incurred by hypoxia.
Keywords: astrocyte; cerebral hypoxia; hypoxia; hypoxia gene module; hypoxia-inducible factors; microglia; neuron; oxygen deprivation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain.J Biol Chem. 1996 Mar 1;271(9):5025-32. doi: 10.1074/jbc.271.9.5025. J Biol Chem. 1996. PMID: 8617779
-
MCT4-mediated expression of EAAT1 is involved in the resistance to hypoxia injury in astrocyte-neuron co-cultures.Neurochem Res. 2015 Apr;40(4):818-28. doi: 10.1007/s11064-015-1532-2. Epub 2015 Feb 3. Neurochem Res. 2015. PMID: 25645447
-
Pericyte, but not astrocyte, hypoxia inducible factor-1 (HIF-1) drives hypoxia-induced vascular permeability in vivo.Fluids Barriers CNS. 2022 Jan 15;19(1):6. doi: 10.1186/s12987-021-00302-y. Fluids Barriers CNS. 2022. PMID: 35033138 Free PMC article.
-
An Overview on the Differential Interplay Among Neurons-Astrocytes-Microglia in CA1 and CA3 Hippocampus in Hypoxia/Ischemia.Front Cell Neurosci. 2020 Nov 11;14:585833. doi: 10.3389/fncel.2020.585833. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33262692 Free PMC article. Review.
-
Lipid Droplets: A Key Cellular Organelle Associated with Cancer Cell Survival under Normoxia and Hypoxia.Int J Mol Sci. 2016 Aug 31;17(9):1430. doi: 10.3390/ijms17091430. Int J Mol Sci. 2016. PMID: 27589734 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

