Identification of an IGHV3-53-Encoded RBD-Targeting Cross-Neutralizing Antibody from an Early COVID-19 Convalescent
- PMID: 38668227
- PMCID: PMC11054858
- DOI: 10.3390/pathogens13040272
Identification of an IGHV3-53-Encoded RBD-Targeting Cross-Neutralizing Antibody from an Early COVID-19 Convalescent
Abstract
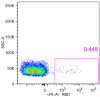
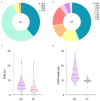
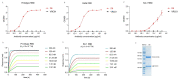
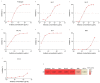
Since November 2021, Omicron has emerged as the dominant severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant, and its sublineages continue to appear one after another, significantly reducing the effectiveness of existing therapeutic neutralizing antibodies (NAbs). It is urgent to develop effective NAbs against circulating Omicron variants. Here, we isolated receptor binding domain (RBD)-specific single memory B cells via flow cytometry from a COVID-19 convalescent. The antibody variable region genes of the heavy chain (VHs) and light chain (VLs) were amplified and cloned into expression vectors. After antibody expression, ELISA screening and neutralizing activity detection, we obtained an IGHV3-53-encoded RBD-targeting cross-neutralizing antibody D6, whose VL originated from the IGKV1-9*01 germlines. D6 could potently neutralize circulating Omicron variants (BA.1, BA.2, BA.4/5 and BF.7), with IC50 values of less than 0.04 μg/mL, and the neutralizing ability against XBB was reduced but still effective. The KD values of D6 binding with RBD of the prototype and BA.1 were both less than 1.0 × 10-12 M. The protein structure of the D6-RBD model indicates that D6 interacts with the RBD external subdomain and belongs to the RBD-1 community. The sufficient contact and deep interaction of D6 HCDR3 and LCDR3 with RBD may be the crucial reason for its cross-neutralizing activity. The sorting and analysis of mAb D6 will provide important information for the development of anti-COVID-19 reagents.
Keywords: Omicron subvariants; RBD; SARS-CoV-2; neutralizing antibody.
Conflict of interest statement
No potential conflicts of interest are reported by the authors.
Figures


Similar articles
-
Pre-Omicron Vaccine Breakthrough Infection Induces Superior Cross-Neutralization against SARS-CoV-2 Omicron BA.1 Compared to Infection Alone.Int J Mol Sci. 2022 Jul 12;23(14):7675. doi: 10.3390/ijms23147675. Int J Mol Sci. 2022. PMID: 35887023 Free PMC article.
-
Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.Proc Natl Acad Sci U S A. 2023 Dec 26;120(52):e2314193120. doi: 10.1073/pnas.2314193120. Epub 2023 Dec 18. Proc Natl Acad Sci U S A. 2023. PMID: 38109549 Free PMC article.
-
A Bispecific Antibody Targeting RBD and S2 Potently Neutralizes SARS-CoV-2 Omicron and Other Variants of Concern.J Virol. 2022 Aug 24;96(16):e0077522. doi: 10.1128/jvi.00775-22. Epub 2022 Aug 2. J Virol. 2022. PMID: 35916510 Free PMC article.
-
Immune evasion of neutralizing antibodies by SARS-CoV-2 Omicron.Cytokine Growth Factor Rev. 2023 Apr;70:13-25. doi: 10.1016/j.cytogfr.2023.03.001. Epub 2023 Mar 5. Cytokine Growth Factor Rev. 2023. PMID: 36948931 Free PMC article. Review.
-
Antibody evasion associated with the RBD significant mutations in several emerging SARS-CoV-2 variants and its subvariants.Drug Resist Updat. 2023 Nov;71:101008. doi: 10.1016/j.drup.2023.101008. Epub 2023 Sep 22. Drug Resist Updat. 2023. PMID: 37757651 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

