This is a preprint.
De novo design of protein minibinder agonists of TLR3
- PMID: 38659926
- PMCID: PMC11042314
- DOI: 10.1101/2024.04.17.589973
De novo design of protein minibinder agonists of TLR3
Abstract
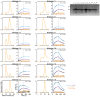
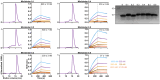
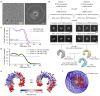
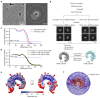
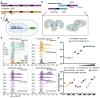
Toll-like Receptor 3 (TLR3) is a pattern recognition receptor that initiates antiviral immune responses upon binding double-stranded RNA (dsRNA). Several nucleic acid-based TLR3 agonists have been explored clinically as vaccine adjuvants in cancer and infectious disease, but present substantial manufacturing and formulation challenges. Here, we use computational protein design to create novel miniproteins that bind to human TLR3 with nanomolar affinities. Cryo-EM structures of two minibinders in complex with TLR3 reveal that they bind the target as designed, although one partially unfolds due to steric competition with a nearby N-linked glycan. Multimeric forms of both minibinders induce NF-κB signaling in TLR3-expressing cell lines, demonstrating that they may have therapeutically relevant biological activity. Our work provides a foundation for the development of specific, stable, and easy-to-formulate protein-based agonists of TLRs and other pattern recognition receptors.
Conflict of interest statement
Declaration of Interests A provisional patent application has been filed by the University of Washington on the TLR3 minibinders described here, listing C.S.A., B.C., and N.P.K. as co-inventors. The King lab has received unrelated sponsored research agreements from Pfizer and GSK. The other authors declare no competing interests.
Figures


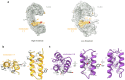






Similar articles
-
Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling.Biochem Biophys Res Commun. 2002 May 24;293(5):1364-9. doi: 10.1016/S0006-291X(02)00380-7. Biochem Biophys Res Commun. 2002. PMID: 12054664
-
The TLR3 signaling complex forms by cooperative receptor dimerization.Proc Natl Acad Sci U S A. 2008 Jan 8;105(1):258-63. doi: 10.1073/pnas.0710779105. Epub 2008 Jan 2. Proc Natl Acad Sci U S A. 2008. PMID: 18172197 Free PMC article.
-
Modulation of double-stranded RNA recognition by the N-terminal histidine-rich region of the human toll-like receptor 3.J Biol Chem. 2008 Aug 15;283(33):22787-94. doi: 10.1074/jbc.M802284200. Epub 2008 Jun 10. J Biol Chem. 2008. PMID: 18544529
-
The toll-like receptor 3:dsRNA signaling complex.Biochim Biophys Acta. 2009 Sep-Oct;1789(9-10):667-74. doi: 10.1016/j.bbagrm.2009.06.005. Epub 2009 Jul 9. Biochim Biophys Acta. 2009. PMID: 19595807 Free PMC article. Review.
-
TLR Agonists as Vaccine Adjuvants Targeting Cancer and Infectious Diseases.Pharmaceutics. 2021 Jan 22;13(2):142. doi: 10.3390/pharmaceutics13020142. Pharmaceutics. 2021. PMID: 33499143 Free PMC article. Review.
References
-
- Taro Kawai S. A. Pathogen recognition with Toll-like receptors. Curr. Opin. Immunol. 17, 338–344 (2005). - PubMed
-
- Medzhitov R., Preston-Hurlburt P. & Janeway C. A. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 (1997). - PubMed
-
- Akira S., Takeda K. & Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680 (2001). - PubMed
-
- Kim H. M. et al. Crystal Structure of the TLR4-MD-2 Complex with Bound Endotoxin Antagonist Eritoran. Cell 130, 906–917 (2007). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
