The involvement of RIPK4 in TNF-α-stimulated IL-6 and IL-8 production by melanoma cells
- PMID: 38656555
- PMCID: PMC11043103
- DOI: 10.1007/s00432-024-05732-3
The involvement of RIPK4 in TNF-α-stimulated IL-6 and IL-8 production by melanoma cells
Abstract
Purpose: The receptor-interacting protein kinase (RIPK4) has an oncogenic function in melanoma, regulates NF-κB and Wnt/β-catenin pathways, and is sensitive to the BRAF inhibitors: vemurafenib and dabrafenib which lead to its decreased level. As its role in melanoma remains not fully understood, we examined the effects of its downregulation on the transcriptomic profile of melanoma.
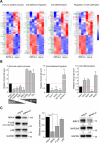
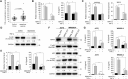
Methods: Applying RNA-seq, we revealed global alterations in the transcriptome of WM266.4 cells with RIPK4 silencing. Functional partners of RIPK4 were evaluated using STRING and GeneMANIA databases. Cells with transient knockdown (via siRNA) and stable knockout (via CRISPR/Cas9) of RIPK4 were stimulated with TNF-α. The expression levels of selected proteins were assessed using Western blot, ELISA, and qPCR.
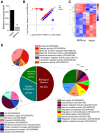
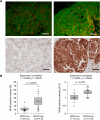
Results: Global analysis of gene expression changes indicates a complex role for RIPK4 in regulating adhesion, migration, proliferation, and inflammatory processes in melanoma cells. Our study highlights potential functional partners of RIPK4 such as BIRC3, TNF-α receptors, and MAP2K6. Data from RIPK4 knockout cells suggest a putative role for RIPK4 in modulating TNF-α-induced production of IL-8 and IL-6 through two distinct signaling pathways-BIRC3/NF-κB and p38/MAPK. Furthermore, increased serum TNF-α levels and the correlation of RIPK4 with NF-κB were revealed in melanoma patients.
Conclusion: These data reveal a complex role for RIPK4 in regulating the immune signaling network in melanoma cells and suggest that this kinase may represent an alternative target for melanoma-targeted adjuvant therapy.
Keywords: BIRC3; Cytokines; Melanoma; RIPK4; RNA-seq; Transcriptome; p-38.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
All authors have declared no conflicts of interest.
Figures

Similar articles
-
RIPK4 downregulation impairs Wnt3A-stimulated invasiveness via Wnt/β-catenin signaling in melanoma cells and tumor growth in vivo.Cell Signal. 2024 Jan;113:110938. doi: 10.1016/j.cellsig.2023.110938. Epub 2023 Oct 21. Cell Signal. 2024. PMID: 37871667
-
Deciphering the Functional Role of RIPK4 in Melanoma.Int J Mol Sci. 2021 Oct 25;22(21):11504. doi: 10.3390/ijms222111504. Int J Mol Sci. 2021. PMID: 34768934 Free PMC article.
-
Vemurafenib and Dabrafenib Downregulates RIPK4 Level.Cancers (Basel). 2023 Feb 1;15(3):918. doi: 10.3390/cancers15030918. Cancers (Basel). 2023. PMID: 36765875 Free PMC article.
-
Signaling pathways for tumor necrosis factor-alpha and interleukin-6 expression in human macrophages exposed to titanium-alloy particulate debris in vitro.J Bone Joint Surg Am. 1999 May;81(5):603-15. doi: 10.2106/00004623-199905000-00002. J Bone Joint Surg Am. 1999. PMID: 10360689
-
Downregulation of RIPK4 Expression Inhibits Epithelial-Mesenchymal Transition in Ovarian Cancer through IL-6.J Immunol Res. 2021 Mar 26;2021:8875450. doi: 10.1155/2021/8875450. eCollection 2021. J Immunol Res. 2021. PMID: 33855091 Free PMC article.
References
-
- Andrews S (2010) FASTQC. A quality control tool for high throughput sequence data, Available online at http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Anghel A-E, Ene C-D, Nicolae I, Budu VA, Constantin C, Neagu M. Interleukin 8-major player in cutaneous melanoma metastatic process. Romanian Biotechnol Lett. 2015;20(6):10911.
-
- Arasu UT, Deen AJ, Pasonen-Seppänen S, Heikkinen S, Lalowski M, Kärnä R, Härkönen K, Mäkinen P, Lázaro-Ibáñez E, Siljander PRM, Oikari S, Levonen AL, Rilla K. HAS3-induced extracellular vesicles from melanoma cells stimulate IHH mediated c-Myc upregulation via the hedgehog signaling pathway in target cells. Cell Mol Life Sci. 2020;77(20):4093–4115. doi: 10.1007/S00018-019-03399-5. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

