Identification of a potent PCNA-p15-interaction inhibitor by autodisplay-based peptide library screening
- PMID: 38646975
- PMCID: PMC11033925
- DOI: 10.1111/1751-7915.14471
Identification of a potent PCNA-p15-interaction inhibitor by autodisplay-based peptide library screening
Abstract
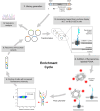
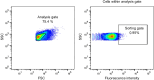
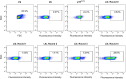
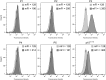
Proliferating cell nuclear antigen (PCNA) is an essential factor for DNA metabolism. The influence of PCNA on DNA replication and repair, combined with the high expression rate of PCNA in various tumours renders PCNA a promising target for cancer therapy. In this context, an autodisplay-based screening method was developed to identify peptidic PCNA interaction inhibitors. A 12-mer randomized peptide library consisting of 2.54 × 106 colony-forming units was constructed and displayed at the surface of Escherichia coli BL21 (DE3) cells by autodisplay. Cells exhibiting an enhanced binding to fluorescent mScarlet-I-PCNA were enriched in four sorting rounds by flow cytometry. This led to the discovery of five peptide variants with affinity to mScarlet-I-PCNA. Among these, P3 (TCPLRWITHDHP) exhibited the highest binding signal. Subsequent flow cytometric analysis revealed a dissociation constant of 0.62 μM for PCNA-P3 interaction. Furthermore, the inhibition of PCNA interactions was investigated using p15, a PIP-box containing protein involved in DNA replication and repair. P3 inhibited the PCNA-p1551-70 interaction with a half maximal inhibitory activity of 16.2 μM, characterizing P3 as a potent inhibitor of the PCNA-p15 interaction.
© 2024 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
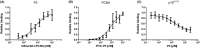
Similar articles
-
Structure of p15(PAF)-PCNA complex and implications for clamp sliding during DNA replication and repair.Nat Commun. 2015 Mar 12;6:6439. doi: 10.1038/ncomms7439. Nat Commun. 2015. PMID: 25762514
-
Unlocking the PIP-box: A peptide library reveals interactions that drive high-affinity binding to human PCNA.J Biol Chem. 2021 Jan-Jun;296:100773. doi: 10.1016/j.jbc.2021.100773. Epub 2021 May 11. J Biol Chem. 2021. PMID: 33984330 Free PMC article.
-
A novel PCNA-binding motif identified by the panning of a random peptide display library.Biochemistry. 2001 Apr 10;40(14):4512-20. doi: 10.1021/bi010103+. Biochemistry. 2001. PMID: 11284708
-
Human PCNA Structure, Function and Interactions.Biomolecules. 2020 Apr 8;10(4):570. doi: 10.3390/biom10040570. Biomolecules. 2020. PMID: 32276417 Free PMC article. Review.
-
Targeting PCNA with Peptide Mimetics for Therapeutic Purposes.Chembiochem. 2020 Feb 17;21(4):442-450. doi: 10.1002/cbic.201900275. Epub 2019 Oct 15. Chembiochem. 2020. PMID: 31247123 Review.
References
-
- Bessette, P.H. , Rice, J.J. & Daugherty, P.S. (2004) Rapid isolation of high‐affinity protein binding peptides using bacterial display. Protein Engineering, Design and Selection, 17, 731–739. - PubMed
-
- Cardano, M. , Tribioli, C. & Prosperi, E. (2020) Targeting proliferating cell nuclear antigen (PCNA) as an effective strategy to inhibit tumor cell proliferation. Current Cancer Drug Targets, 20, 240–252. - PubMed
-
- Daugherty, P.S. , Chen, G. , Olsen, M.J. , Iverson, B.L. & Georgiou, G. (1998) Antibody affinity maturation using bacterial surface display. Protein Engineering, 11, 825–832. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

