Bacterial peptidoglycan acts as a digestive signal mediating host adaptation to diverse food resources in C. elegans
- PMID: 38627398
- PMCID: PMC11021419
- DOI: 10.1038/s41467-024-47530-y
Bacterial peptidoglycan acts as a digestive signal mediating host adaptation to diverse food resources in C. elegans
Abstract
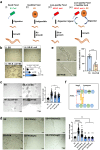
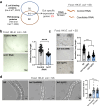
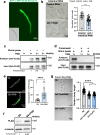
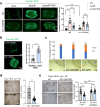
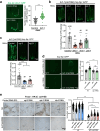
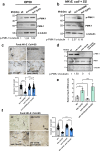
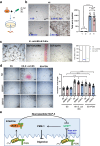
Food availability and usage is a major adaptive force for the successful survival of animals in nature, yet little is known about the specific signals that activate the host digestive system to allow for the consumption of varied foods. Here, by using a food digestion system in C. elegans, we discover that bacterial peptidoglycan (PGN) is a unique food signal that activates animals to digest inedible food. We identified that a glycosylated protein, Bacterial Colonization Factor-1 (BCF-1), in the gut interacts with bacterial PGN, leading to the inhibition of the mitochondrial unfolded protein response (UPRmt) by regulating the release of Neuropeptide-Like Protein (NLP-3). Interestingly, activating UPRmt was found to hinder food digestion, which depends on the innate immune p38 MAPK/PMK-1 pathway. Conversely, inhibiting PMK-1 was able to alleviate digestion defects in bcf-1 mutants. Furthermore, we demonstrate that animals with digestion defects experience reduced natural adaptation capabilities. This study reveals that PGN-BCF-1 interaction acts as "good-food signal" to promote food digestion and animal growth, which facilitates adaptation of the host animals by increasing ability to consume a wide range of foods in their natural environment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Genedrive kit for detecting single nucleotide polymorphism m.1555A>G in neonates and their mothers: a systematic review and cost-effectiveness analysis.Health Technol Assess. 2024 Oct;28(75):1-75. doi: 10.3310/TGAC4201. Health Technol Assess. 2024. PMID: 39487741 Free PMC article.
-
Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article.
-
Distinct functions of three Wnt proteins control mirror-symmetric organogenesis in the C. elegans gonad.Elife. 2024 Nov 1;13:e103035. doi: 10.7554/eLife.103035. Elife. 2024. PMID: 39485276 Free PMC article.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
The hidden base of the iceberg: gut peptidoglycome dynamics is foundational to its influence on the host.Gut Microbes. 2024 Jan-Dec;16(1):2395099. doi: 10.1080/19490976.2024.2395099. Epub 2024 Sep 6. Gut Microbes. 2024. PMID: 39239828 Free PMC article. Review.
References
-
- Freckman DW. Bacterivorous nematodes and organic-matter decomposition. Agr. Ecosyst. Environ. 1988;24:195–217. doi: 10.1016/0167-8809(88)90066-7. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

