Hammerhead-type FXR agonists induce an enhancer RNA Fincor that ameliorates nonalcoholic steatohepatitis in mice
- PMID: 38619504
- PMCID: PMC11018349
- DOI: 10.7554/eLife.91438
Hammerhead-type FXR agonists induce an enhancer RNA Fincor that ameliorates nonalcoholic steatohepatitis in mice
Abstract
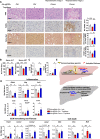
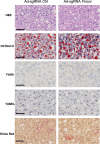
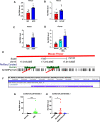
The nuclear receptor, farnesoid X receptor (FXR/NR1H4), is increasingly recognized as a promising drug target for metabolic diseases, including nonalcoholic steatohepatitis (NASH). Protein-coding genes regulated by FXR are well known, but whether FXR also acts through regulation of long non-coding RNAs (lncRNAs), which vastly outnumber protein-coding genes, remains unknown. Utilizing RNA-seq and global run-on sequencing (GRO-seq) analyses in mouse liver, we found that FXR activation affects the expression of many RNA transcripts from chromatin regions bearing enhancer features. Among these we discovered a previously unannotated liver-enriched enhancer-derived lncRNA (eRNA), termed FXR-induced non-coding RNA (Fincor). We show that Fincor is specifically induced by the hammerhead-type FXR agonists, including GW4064 and tropifexor. CRISPR/Cas9-mediated liver-specific knockdown of Fincor in dietary NASH mice reduced the beneficial effects of tropifexor, an FXR agonist currently in clinical trials for NASH and primary biliary cholangitis (PBC), indicating that amelioration of liver fibrosis and inflammation in NASH treatment by tropifexor is mediated in part by Fincor. Overall, our findings highlight that pharmacological activation of FXR by hammerhead-type agonists induces a novel eRNA, Fincor, contributing to the amelioration of NASH in mice. Fincor may represent a new drug target for addressing metabolic disorders, including NASH.
Keywords: FXR; NASH; enhancer RNA; human; medicine; mouse; tropifexor.
Plain language summary
Non-alcoholic steatohepatitis, also known as NASH, is a severe condition whereby fat deposits around the liver lead to inflammation, swelling, scarring and lasting damage to the organ. Despite being one of the leading causes of liver-related deaths worldwide, the disease has no approved treatment. A protein known as Farnesoid X receptor (or FXR) is increasingly being recognized as a promising drug target for non-alcoholic steatohepatitis. Once activated, FXR helps to regulate the activity of DNA regions which are coding for proteins important for liver health. However, less is known about how FXR may act on non-coding regions, the DNA sequences that do not generate proteins but can be transcribed into RNA molecules with important biological roles. In response, Chen et al. investigated whether FXR activation of non-coding RNAs could be linked to the clinical benefits of hammerhead FXR agonists, a type of synthetic compounds that activates this receptor. To do so, genetic analyses of mouse livers were performed to identify non-coding RNAs generated when FXR was activated by the agonist. These experiments revealed that agonist-activated FXR induced a range of non-coding RNAs transcribed from DNA sequences known as enhancers, which help to regulate gene expression. In particular, hammerhead FXR agonists led to the production of a liver-specific enhancer RNA called Fincor. Additional experiments using tropifexor, a hammerhead FXR agonist currently into clinical trials, showed that this investigational new drug had reduced benefits in a mouse model of non-alcoholic steatohepatitis with low Fincor levels. This suggested that this enhancer RNA may play a key role in mediating the clinical benefits of hammerhead FXR agonists, encouraging further research into its role and therapeutic value.
© 2024, Chen, Wang et al.
Conflict of interest statement
JC, RW, FX, HS, BK, WL, JK No competing interests declared
Figures


Update of
-
Hammerhead-type FXR agonists induce an eRNA FincoR that ameliorates nonalcoholic steatohepatitis in mice.bioRxiv [Preprint]. 2024 Feb 8:2023.11.20.567833. doi: 10.1101/2023.11.20.567833. bioRxiv. 2024. Update in: Elife. 2024 Apr 15;13:RP91438. doi: 10.7554/eLife.91438. PMID: 38045226 Free PMC article. Updated. Preprint.
Similar articles
-
Hammerhead-type FXR agonists induce an eRNA FincoR that ameliorates nonalcoholic steatohepatitis in mice.bioRxiv [Preprint]. 2024 Feb 8:2023.11.20.567833. doi: 10.1101/2023.11.20.567833. bioRxiv. 2024. Update in: Elife. 2024 Apr 15;13:RP91438. doi: 10.7554/eLife.91438. PMID: 38045226 Free PMC article. Updated. Preprint.
-
Tropifexor-Mediated Abrogation of Steatohepatitis and Fibrosis Is Associated With the Antioxidative Gene Expression Profile in Rodents.Hepatol Commun. 2019 May 17;3(8):1085-1097. doi: 10.1002/hep4.1368. eCollection 2019 Aug. Hepatol Commun. 2019. PMID: 31388629 Free PMC article.
-
Bile acid modulators for the treatment of nonalcoholic steatohepatitis (NASH).Expert Opin Investig Drugs. 2020 Jun;29(6):623-632. doi: 10.1080/13543784.2020.1763302. Epub 2020 Jun 19. Expert Opin Investig Drugs. 2020. PMID: 32552182 Review.
-
Discovery of Tropifexor (LJN452), a Highly Potent Non-bile Acid FXR Agonist for the Treatment of Cholestatic Liver Diseases and Nonalcoholic Steatohepatitis (NASH).J Med Chem. 2017 Dec 28;60(24):9960-9973. doi: 10.1021/acs.jmedchem.7b00907. Epub 2017 Dec 8. J Med Chem. 2017. PMID: 29148806
-
Obeticholic acid-a new therapy in PBC and NASH.Br Med Bull. 2020 May 15;133(1):95-104. doi: 10.1093/bmb/ldaa006. Br Med Bull. 2020. PMID: 32282030 Review.
References
-
- Abel U, Schlüter T, Schulz A, Hambruch E, Steeneck C, Hornberger M, Hoffmann T, Perović-Ottstadt S, Kinzel O, Burnet M, Deuschle U, Kremoser C. Synthesis and pharmacological validation of a novel series of non-steroidal FXR agonists. Bioorganic & Medicinal Chemistry Letters. 2010;20:4911–4917. doi: 10.1016/j.bmcl.2010.06.084. - DOI - PubMed
-
- Brocker CN, Kim D, Melia T, Karri K, Velenosi TJ, Takahashi S, Aibara D, Bonzo JA, Levi M, Waxman DJ, Gonzalez FJ. Long non-coding RNA Gm15441 attenuates hepatic inflammasome activation in response to PPARA agonism and fasting. Nature Communications. 2020;11:5847. doi: 10.1038/s41467-020-19554-7. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

