This is a preprint.
A specific and portable gene expression program underlies antigen archiving by lymphatic endothelial cells
- PMID: 38617225
- PMCID: PMC11014631
- DOI: 10.1101/2024.04.01.587647
A specific and portable gene expression program underlies antigen archiving by lymphatic endothelial cells
Abstract
Antigens from protein subunit vaccination traffic from the tissue to the draining lymph node, either passively via the lymph or carried by dendritic cells at the local injection site. Lymph node (LN) lymphatic endothelial cells (LEC) actively acquire and archive foreign antigens, and archived antigen can be released during subsequent inflammatory stimulus to improve immune responses. Here, we answer questions about how LECs achieve durable antigen archiving and whether there are transcriptional signatures associated with LECs containing high levels of antigen. We used single cell sequencing in dissociated LN tissue to quantify antigen levels in LEC and dendritic cell populations at multiple timepoints after immunization, and used machine learning to define a unique transcriptional program within archiving LECs that can predict LEC archiving capacity in independent data sets. Finally, we validated this modeling, showing we could predict antigen archiving from a transcriptional dataset of CHIKV infected mice and demonstrated in vivo the accuracy of our prediction. Collectively, our findings establish a unique transcriptional program in LECs that promotes antigen archiving that can be translated to other systems.
Keywords: Chikungunya virus; antigen archiving; dendritic cell; fibroblastic reticular cell; gene expression program; immunization; lymph node; lymph node stromal cell; lymphatic endothelial cell.
Conflict of interest statement
CONFLICT OF INTEREST We declare no competing interest.
Figures



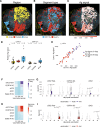
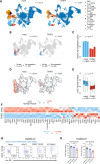
Similar articles
-
Immunization-induced antigen archiving enhances local memory CD8+ T cell responses following an unrelated viral infection.NPJ Vaccines. 2024 Mar 21;9(1):66. doi: 10.1038/s41541-024-00856-6. NPJ Vaccines. 2024. PMID: 38514656 Free PMC article.
-
Chikungunya virus infection disrupts lymph node lymphatic endothelial cell composition and function via MARCO.JCI Insight. 2024 Jan 9;9(4):e176537. doi: 10.1172/jci.insight.176537. JCI Insight. 2024. PMID: 38194268 Free PMC article.
-
Molecular tracking devices quantify antigen distribution and archiving in the murine lymph node.Elife. 2021 Apr 12;10:e62781. doi: 10.7554/eLife.62781. Elife. 2021. PMID: 33843587 Free PMC article.
-
Antigen archiving by lymph node stroma: A novel function for the lymphatic endothelium.Eur J Immunol. 2015 Oct;45(10):2721-9. doi: 10.1002/eji.201545739. Epub 2015 Sep 10. Eur J Immunol. 2015. PMID: 26278423 Free PMC article. Review.
-
Lymph Node Lymphatic Endothelial Cell Expansion and Contraction and the Programming of the Immune Response.Front Immunol. 2019 Jan 25;10:36. doi: 10.3389/fimmu.2019.00036. eCollection 2019. Front Immunol. 2019. PMID: 30740101 Free PMC article. Review.
References
-
- Takamura S, Roberts AD, Jelley-Gibbs DM, Wittmer ST, Kohlmeier JE, Woodland DL. The route of priming influences the ability of respiratory virus-specific memory CD8+ T cells to be activated by residual antigen. J Exp Med. 2010;207(6):1153–60. doi: jem.20090283 [pii] 10.1084/jem.20090283. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
