Growth-regulated co-occupancy of Mediator and Lsm3 at intronic ribosomal protein genes
- PMID: 38613396
- PMCID: PMC11194063
- DOI: 10.1093/nar/gkae266
Growth-regulated co-occupancy of Mediator and Lsm3 at intronic ribosomal protein genes
Erratum in
-
Correction to "Growth-regulated co-occupancy of Mediator and Lsm3 at intronic ribosomal protein genes".Nucleic Acids Res. 2025 Jan 7;53(1):gkae1258. doi: 10.1093/nar/gkae1258. Nucleic Acids Res. 2025. PMID: 39704121 Free PMC article. No abstract available.
Abstract
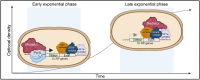
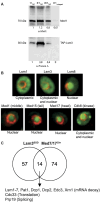
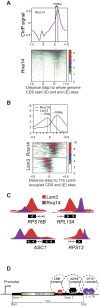
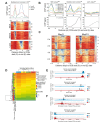
Mediator is a well-known transcriptional co-regulator and serves as an adaptor between gene-specific regulatory proteins and RNA polymerase II. Studies on the chromatin-bound form of Mediator revealed interactions with additional protein complexes involved in various transcription-related processes, such as the Lsm2-8 complex that is part of the spliceosomal U6 small nuclear ribonucleoprotein complex. Here, we employ Chromatin Immunoprecipitation sequencing (ChIP-seq) of chromatin associated with the Lsm3 protein and the Med1 or Med15 Mediator subunits. We identify 86 genes co-occupied by both Lsm3 and Mediator, of which 73 were intron-containing ribosomal protein genes. In logarithmically growing cells, Mediator primarily binds to their promoter regions but also shows a second, less pronounced occupancy at their 3'-exons. During the late exponential phase, we observe a near-complete transition of Mediator from these promoters to a position in their 3'-ends, overlapping the Lsm3 binding sites ∼250 bp downstream of their last intron-exon boundaries. Using an unbiased RNA sequencing approach, we show that transition of Mediator from promoters to the last exon of these genes correlates to reduction of both their messenger RNA levels and splicing ratios, indicating that the Mediator and Lsm complexes cooperate to control growth-regulated expression of intron-containing ribosomal protein genes at the levels of transcription and splicing.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
"I've Spent My Whole Life Striving to Be Normal": Internalized Stigma and Perceived Impact of Diagnosis in Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):423-436. doi: 10.1089/aut.2022.0066. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116050 Free PMC article.
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article. Review.
References
-
- Kim Y.-J., Björklund S., Li Y., Sayre M.H., Kornberg R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994; 77:599–608. - PubMed
-
- Thompson C.M., Koleske A.J., Chao D.M., Young R.A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993; 73:1361–1375. - PubMed
-
- Yudkovsky N., Ranish J.A., Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000; 408:225–229. - PubMed
-
- Chereji R.V., Bharatula V., Elfving N., Blomberg J., Larsson M., Morozov A.V., Broach J.R., Björklund S. Mediator binds to boundaries of chromosomal interaction domains and to proteins involved in DNA looping, RNA metabolism, chromatin remodeling, and actin assembly. Nucleic Acids Res. 2017; 45:8806–8821. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

