A novel inhibitory BAK antibody enables assessment of non-activated BAK in cancer cells
- PMID: 38582955
- PMCID: PMC11164899
- DOI: 10.1038/s41418-024-01289-3
A novel inhibitory BAK antibody enables assessment of non-activated BAK in cancer cells
Abstract
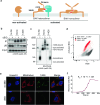
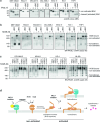
BAX and BAK are pro-apoptotic members of the BCL2 family that are required to permeabilize the mitochondrial outer membrane. The proteins can adopt a non-activated monomeric conformation, or an activated conformation in which the exposed BH3 domain facilitates binding either to a prosurvival protein or to another activated BAK or BAX protein to promote pore formation. Certain cancer cells are proposed to have high levels of activated BAK sequestered by MCL1 or BCLXL, thus priming these cells to undergo apoptosis in response to BH3 mimetic compounds that target MCL1 or BCLXL. Here we report the first antibody, 14G6, that is specific for the non-activated BAK conformer. A crystal structure of 14G6 Fab bound to BAK revealed a binding site encompassing both the α1 helix and α5-α6 hinge regions of BAK, two sites involved in the unfolding of BAK during its activation. In mitochondrial experiments, 14G6 inhibited BAK unfolding triggered by three diverse BAK activators, supporting crucial roles for both α1 dissociation and separation of the core (α2-α5) and latch (α6-α9) regions in BAK activation. 14G6 bound the majority of BAK in several leukaemia cell lines, and binding decreased following treatment with BH3 mimetics, indicating only minor levels of constitutively activated BAK in those cells. In summary, 14G6 provides a new means of assessing BAK status in response to anti-cancer treatments.
© 2024. Crown.
Conflict of interest statement
HPSS, SI, MXS, AWW, KCF, AZW, DL, JMB, RTU, PEC, MSM and RMK are or were employees of WEHI which receives royalties from AbbVie and Genentech from the sale of Venetoclax.
Figures


Similar articles
-
Constitutive BAK activation as a determinant of drug sensitivity in malignant lymphohematopoietic cells.Genes Dev. 2015 Oct 15;29(20):2140-52. doi: 10.1101/gad.267997.115. Genes Dev. 2015. PMID: 26494789 Free PMC article.
-
Assembly of the Bak apoptotic pore: a critical role for the Bak protein α6 helix in the multimerization of homodimers during apoptosis.J Biol Chem. 2013 Sep 6;288(36):26027-26038. doi: 10.1074/jbc.M113.490094. Epub 2013 Jul 26. J Biol Chem. 2013. PMID: 23893415 Free PMC article.
-
In non-transformed cells Bak activates upon loss of anti-apoptotic Bcl-XL and Mcl-1 but in the absence of active BH3-only proteins.Cell Death Dis. 2015 Nov 26;6(11):e1996. doi: 10.1038/cddis.2015.341. Cell Death Dis. 2015. PMID: 26610208 Free PMC article.
-
BCL2 and MCL1 inhibitors for hematologic malignancies.Blood. 2021 Sep 30;138(13):1120-1136. doi: 10.1182/blood.2020006785. Blood. 2021. PMID: 34320168 Review.
-
Physiological and Pharmacological Control of BAK, BAX, and Beyond.Trends Cell Biol. 2016 Dec;26(12):906-917. doi: 10.1016/j.tcb.2016.07.002. Epub 2016 Aug 4. Trends Cell Biol. 2016. PMID: 27498846 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- GNT1113133/Department of Health | National Health and Medical Research Council (NHMRC)
- GNT2009062/Department of Health | National Health and Medical Research Council (NHMRC)
- 2001406/Department of Health | National Health and Medical Research Council (NHMRC)
- 7015-18/Leukemia and Lymphoma Society (Leukemia & Lymphoma Society)
LinkOut - more resources
Full Text Sources
Research Materials

