Cuproptosis-related gene-located DNA methylation in lower-grade glioma: Prognosis and tumor microenvironment
- PMID: 38578883
- PMCID: PMC11307024
- DOI: 10.3233/CBM-230341
Cuproptosis-related gene-located DNA methylation in lower-grade glioma: Prognosis and tumor microenvironment
Abstract
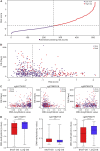
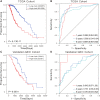
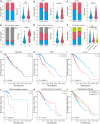
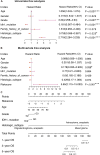
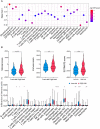
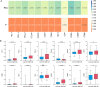
Cuproptosis a novel copper-dependent cell death modality, plays a crucial part in the oncogenesis, progression and prognosis of tumors. However, the relationships among DNA-methylation located in cuproptosis-related genes (CRGs), overall survival (OS) and the tumor microenvironment remain undefined. In this study, we systematically assessed the prognostic value of CRG-located DNA-methylation for lower-grade glioma (LGG). Clinical and molecular data were sourced from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. We employed Cox hazard regression to examine the associations between CRG-located DNA-methylation and OS, leading to the development of a prognostic signature. Kaplan-Meier survival and time-dependent receiver operating characteristic (ROC) analyses were utilized to gauge the accuracy of the signature. Gene Set Enrichment Analysis (GSEA) was applied to uncover potential biological functions of differentially expressed genes between high- and low-risk groups. A three CRG-located DNA-methylation prognostic signature was established based on TCGA database and validated in GEO dataset. The 1-year, 3-year, and 5-year area under the curve (AUC) of ROC curves in the TCGA dataset were 0.884, 0.888, and 0.859 while those in the GEO dataset were 0.943, 0.761 and 0.725, respectively. Cox-regression-analyses revealed the risk signature as an independent risk factor for LGG patients. Immunogenomic profiling suggested that the signature was associated with immune infiltration level and immune checkpoints. Functional enrichment analysis indicated differential enrichment in cell differentiation in the hindbrain, ECM receptor interactions, glycolysis and reactive oxygen species pathway across different groups. We developed and verified a novel CRG-located DNA-methylation signature to predict the prognosis in LGG patients. Our findings emphasize the potential clinical implications of CRG-located DNA-methylation indicating that it may serve as a promising therapeutic target for LGG patients.
Keywords: DNA-methylation; Lower-grade glioma; cuproptosis; immune checkpoint inhibitors; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Identification of a novel cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas.Front Immunol. 2022 Aug 15;13:933973. doi: 10.3389/fimmu.2022.933973. eCollection 2022. Front Immunol. 2022. PMID: 36045691 Free PMC article. Review.
-
Identification of cuproptosis-related subtypes, construction of a prognosis model, and tumor microenvironment landscape in gastric cancer.Front Immunol. 2022 Nov 21;13:1056932. doi: 10.3389/fimmu.2022.1056932. eCollection 2022. Front Immunol. 2022. PMID: 36479114 Free PMC article.
-
A cuproptosis-based prognostic model for predicting survival in low-grade glioma.Aging (Albany NY). 2024 May 9;16(10):8697-8716. doi: 10.18632/aging.205834. Epub 2024 May 9. Aging (Albany NY). 2024. PMID: 38738989 Free PMC article.
-
Cuproptosis-Related Signature Predicts the Prognosis, Tumor Microenvironment, and Drug Sensitivity of Hepatocellular Carcinoma.J Immunol Res. 2022 Nov 16;2022:3393027. doi: 10.1155/2022/3393027. eCollection 2022. J Immunol Res. 2022. PMID: 36438201 Free PMC article.
-
Radiomic Prediction of CCND1 Expression Levels and Prognosis in Low-grade Glioma Based on Magnetic Resonance Imaging.Acad Radiol. 2024 Nov;31(11):4595-4610. doi: 10.1016/j.acra.2024.03.031. Epub 2024 Jun 1. Acad Radiol. 2024. PMID: 38824087 Review.
Cited by
-
Gene Expression Regulation and the Signal Transduction of Programmed Cell Death.Curr Issues Mol Biol. 2024 Sep 16;46(9):10264-10298. doi: 10.3390/cimb46090612. Curr Issues Mol Biol. 2024. PMID: 39329964 Free PMC article. Review.
References
-
- Song L.R. et al., Identification and validation of a 21-mRNA prognostic signature in diffuse lower-grade gliomas, J Neurooncol 146(1) (2020), 207–217. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

