DoUBLing up: ubiquitin and ubiquitin-like proteases in genome stability
- PMID: 38572758
- PMCID: PMC11088880
- DOI: 10.1042/BCJ20230284
DoUBLing up: ubiquitin and ubiquitin-like proteases in genome stability
Abstract
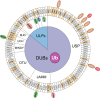
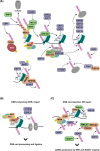
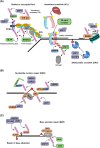
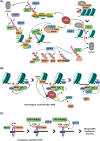
Maintaining stability of the genome requires dedicated DNA repair and signalling processes that are essential for the faithful duplication and propagation of chromosomes. These DNA damage response (DDR) mechanisms counteract the potentially mutagenic impact of daily genotoxic stresses from both exogenous and endogenous sources. Inherent to these DNA repair pathways is the activity of protein factors that instigate repair processes in response to DNA lesions. The regulation, coordination, and orchestration of these DDR factors is carried out, in a large part, by post-translational modifications, such as phosphorylation, ubiquitylation, and modification with ubiquitin-like proteins (UBLs). The importance of ubiquitylation and UBLylation with SUMO in DNA repair is well established, with the modified targets and downstream signalling consequences relatively well characterised. However, the role of dedicated erasers for ubiquitin and UBLs, known as deubiquitylases (DUBs) and ubiquitin-like proteases (ULPs) respectively, in genome stability is less well established, particularly for emerging UBLs such as ISG15 and UFM1. In this review, we provide an overview of the known regulatory roles and mechanisms of DUBs and ULPs involved in genome stability pathways. Expanding our understanding of the molecular agents and mechanisms underlying the removal of ubiquitin and UBL modifications will be fundamental for progressing our knowledge of the DDR and likely provide new therapeutic avenues for relevant human diseases, such as cancer.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
Similar articles
-
Ubiquitin-like proteins in the DNA damage response: the next generation.Essays Biochem. 2020 Oct 26;64(5):737-752. doi: 10.1042/EBC20190095. Essays Biochem. 2020. PMID: 32451552 Review.
-
Chemical biology tools to study Deubiquitinases and Ubl proteases.Semin Cell Dev Biol. 2022 Dec;132:86-96. doi: 10.1016/j.semcdb.2022.02.006. Epub 2022 Feb 22. Semin Cell Dev Biol. 2022. PMID: 35216867 Review.
-
Ubiquitin-like modifications in the DNA damage response.Mutat Res. 2017 Oct;803-805:56-75. doi: 10.1016/j.mrfmmm.2017.07.001. Epub 2017 Jul 11. Mutat Res. 2017. PMID: 28734548 Review.
-
Ubiquitylation, neddylation and the DNA damage response.Open Biol. 2015 Apr;5(4):150018. doi: 10.1098/rsob.150018. Open Biol. 2015. PMID: 25833379 Free PMC article. Review.
-
Tools for Decoding Ubiquitin Signaling in DNA Repair.Front Cell Dev Biol. 2021 Dec 7;9:760226. doi: 10.3389/fcell.2021.760226. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34950659 Free PMC article. Review.
Cited by
-
SUMO and the DNA damage response.Biochem Soc Trans. 2024 Apr 24;52(2):773-792. doi: 10.1042/BST20230862. Biochem Soc Trans. 2024. PMID: 38629643 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

