Identification of molecular subtypes and a prognostic signature based on m6A/m5C/m1A-related genes in lung adenocarcinoma
- PMID: 38555384
- PMCID: PMC10981664
- DOI: 10.1038/s41598-024-57910-5
Identification of molecular subtypes and a prognostic signature based on m6A/m5C/m1A-related genes in lung adenocarcinoma
Abstract
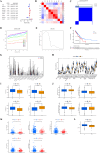
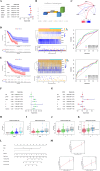
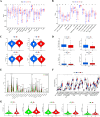
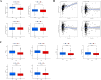
Lung cancer, specifically the histological subtype lung adenocarcinoma (LUAD), has the highest global occurrence and fatality rate. Extensive research has indicated that RNA alterations encompassing m6A, m5C, and m1A contribute actively to tumorigenesis, drug resistance, and immunotherapy responses in LUAD. Nevertheless, the absence of a dependable predictive model based on m6A/m5C/m1A-associated genes hinders accurately predicting the prognosis of patients diagnosed with LUAD. In this study, we collected patient data from The Cancer Genome Atlas (TCGA) and identified genes related to m6A/m5C/m1A modifications using the GeneCards database. The "ConsensusClusterPlus" R package was used to produce molecular subtypes by utilizing genes relevant to m6A/m5C/m1A identified through differential expression and univariate Cox analyses. An independent prognostic factor was identified by constructing a prognostic signature comprising six genes (SNHG12, PABPC1, IGF2BP1, FOXM1, CBFA2T3, and CASC8). Poor overall survival and elevated expression of human leukocyte antigens and immune checkpoints were correlated with higher risk scores. We examined the associations between the sets of genes regulated by m6A/m5C/m1A and the risk model, as well as the immune cell infiltration, using algorithms such as ESTIMATE, CIBERSORT, TIMER, ssGSEA, and exclusion (TIDE). Moreover, we compared tumor stemness indices (TSIs) by considering the molecular subtypes related to m6A/m5C/m1A and risk signatures. Analyses were performed based on the risk signature, including stratification, somatic mutation analysis, nomogram construction, chemotherapeutic response prediction, and small-molecule drug prediction. In summary, we developed a prognostic signature consisting of six genes that have the potential for prognostication in patients with LUAD and the design of personalized treatments that could provide new versions of personalized management for these patients.
Keywords: Lung adenocarcinoma; Molecule subtypes; Signature; m1A; m5C; m6A.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The role of m5C RNA modification in cancer development and therapy.Heliyon. 2024 Sep 27;10(19):e38660. doi: 10.1016/j.heliyon.2024.e38660. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39444404 Free PMC article. Review.
-
Development of m6A/m5C/m1A regulated lncRNA signature for prognostic prediction, personalized immune intervention and drug selection in LUAD.J Cell Mol Med. 2024 Apr;28(8):e18282. doi: 10.1111/jcmm.18282. J Cell Mol Med. 2024. PMID: 38647237 Free PMC article.
-
Construction of a prognostic model for lung adenocarcinoma based on m6A/m5C/m1A genes.Hum Mol Genet. 2024 Mar 20;33(7):563-582. doi: 10.1093/hmg/ddad208. Hum Mol Genet. 2024. PMID: 38142284
-
Exploration and validation of a combined Hypoxia and m6A/m5C/m1A regulated gene signature for prognosis prediction of liver cancer.BMC Genomics. 2023 Dec 14;24(1):776. doi: 10.1186/s12864-023-09876-3. BMC Genomics. 2023. PMID: 38097948 Free PMC article.
-
A review of current developments in RNA modifications in lung cancer.Cancer Cell Int. 2024 Oct 25;24(1):347. doi: 10.1186/s12935-024-03528-6. Cancer Cell Int. 2024. PMID: 39456034 Free PMC article. Review.
Cited by
-
The role of m5C RNA modification in cancer development and therapy.Heliyon. 2024 Sep 27;10(19):e38660. doi: 10.1016/j.heliyon.2024.e38660. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39444404 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- 2024Y221/Scientific Research Fund Project of Education Department of Yunnan Province
- 2024Y221/Scientific Research Fund Project of Education Department of Yunnan Province
- CXTD202210/Science and Technology Innovation Team for Precision Pathological Diagnosis of Lung Malignant Tumours at Kunming Medical University
- CXTD202210/Science and Technology Innovation Team for Precision Pathological Diagnosis of Lung Malignant Tumours at Kunming Medical University
- 82360523/the Regional Fund Project of the National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

