TNFR1 mediates heterogeneity in single-cell NF-κB activation
- PMID: 38551009
- PMCID: PMC10973173
- DOI: 10.1016/j.isci.2024.109486
TNFR1 mediates heterogeneity in single-cell NF-κB activation
Abstract
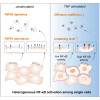
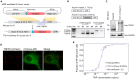
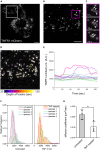
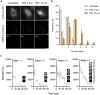
Nuclear factor kappa B (NF-κB) is a key regulator in immune signaling and is known to exhibit a digital activation pattern. Yet the molecular basis underlying the heterogeneity in NF-κB activation at single-cell level is not entirely understood. Here, we show that NF-κB activation in single cells is largely regulated by intrinsic differences at the receptor level. Using the genome editing and time-lapse imaging, we directly characterize endogenous TNFR1 dynamics and NF-κB activation from the same single cells. Total internal reflection fluorescence (TIRF) microscopy shows that endogenous TNFR1 forms pre-ligand clusters in the resting cells. Upon tumor necrosis factor (TNF) stimulation, the diffusion coefficient of membrane TNFR1 was significantly decreased and a substantial level of TNFR1 undergoes oligomerization to form trimers and hexamers. Moreover, multi-color cell imaging reveals that both digital and graded information processing regulate NF-κB activation across different TNFR1 expression levels. Our results indicate that single-cell NF-κB activation potential strongly correlates with its TNFR1 characteristics.
Keywords: Biological sciences; Cell biology; Molecular biology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
HBx-induced hepatic steatosis and apoptosis are regulated by TNFR1- and NF-kappaB-dependent pathways.J Mol Biol. 2010 Apr 9;397(4):917-31. doi: 10.1016/j.jmb.2010.02.016. Epub 2010 Feb 13. J Mol Biol. 2010. PMID: 20156456
-
Single-molecule imaging reveals the oligomeric state of functional TNFα-induced plasma membrane TNFR1 clusters in cells.Sci Signal. 2020 Jan 14;13(614):eaax5647. doi: 10.1126/scisignal.aax5647. Sci Signal. 2020. PMID: 31937565
-
Microglia-derived TNF-α mediates Müller cell activation by activating the TNFR1-NF-κB pathway.Exp Eye Res. 2022 Jan;214:108852. doi: 10.1016/j.exer.2021.108852. Epub 2021 Nov 18. Exp Eye Res. 2022. PMID: 34801535
-
TRAF2 multitasking in TNF receptor-induced signaling to NF-κB, MAP kinases and cell death.Biochem Pharmacol. 2016 Sep 15;116:1-10. doi: 10.1016/j.bcp.2016.03.009. Epub 2016 Mar 16. Biochem Pharmacol. 2016. PMID: 26993379 Review.
-
TNFR1-activated NF-κB signal transduction: regulation by the ubiquitin/proteasome system.Curr Opin Chem Biol. 2014 Dec;23:71-7. doi: 10.1016/j.cbpa.2014.10.011. Curr Opin Chem Biol. 2014. PMID: 25461388 Review.

