voyAGEr, a free web interface for the analysis of age-related gene expression alterations in human tissues
- PMID: 38546191
- PMCID: PMC10977969
- DOI: 10.7554/eLife.88623
voyAGEr, a free web interface for the analysis of age-related gene expression alterations in human tissues
Abstract
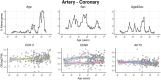
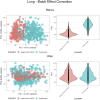
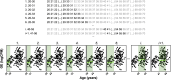
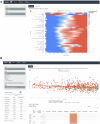
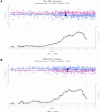
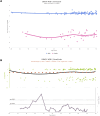
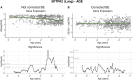
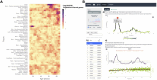
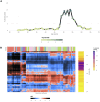
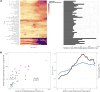
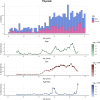
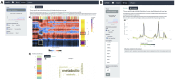
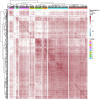
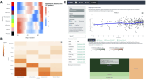
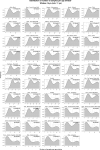
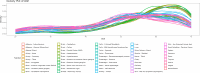
We herein introduce voyAGEr, an online graphical interface to explore age-related gene expression alterations in 49 human tissues. voyAGEr offers a visualisation and statistical toolkit for the finding and functional exploration of sex- and tissue-specific transcriptomic changes with age. In its conception, we developed a novel bioinformatics pipeline leveraging RNA sequencing data, from the GTEx project, encompassing more than 900 individuals. voyAGEr reveals transcriptomic signatures of the known asynchronous ageing between tissues, allowing the observation of tissue-specific age periods of major transcriptional changes, associated with alterations in different biological pathways, cellular composition, and disease conditions. Notably, voyAGEr was created to assist researchers with no expertise in bioinformatics, providing a supportive framework for elaborating, testing and refining their hypotheses on the molecular nature of human ageing and its association with pathologies, thereby also aiding in the discovery of novel therapeutic targets. voyAGEr is freely available at https://compbio.imm.medicina.ulisboa.pt/app/voyAGEr.
Keywords: ageing; chromosomes; computational biology; gene expression; gene expression profiling; human; systems biology; transcriptome.
© 2023, Schneider, Martins-Silva, Kaizeler et al.
Conflict of interest statement
AS, RM, AK, NS, NB No competing interests declared
Figures





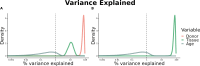
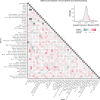
Update of
- doi: 10.1101/2022.12.22.521681
- doi: 10.7554/eLife.88623.1
- doi: 10.7554/eLife.88623.2
Similar articles
-
RNA-seq based transcriptomic map reveals new insights into mouse salivary gland development and maturation.BMC Genomics. 2016 Nov 16;17(1):923. doi: 10.1186/s12864-016-3228-7. BMC Genomics. 2016. PMID: 27852218 Free PMC article.
-
Eye in a Disk: eyeIntegration Human Pan-Eye and Body Transcriptome Database Version 1.0.Invest Ophthalmol Vis Sci. 2019 Jul 1;60(8):3236-3246. doi: 10.1167/iovs.19-27106. Invest Ophthalmol Vis Sci. 2019. PMID: 31343654 Free PMC article.
-
RNA CoMPASS: a dual approach for pathogen and host transcriptome analysis of RNA-seq datasets.PLoS One. 2014 Feb 25;9(2):e89445. doi: 10.1371/journal.pone.0089445. eCollection 2014. PLoS One. 2014. PMID: 24586784 Free PMC article.
-
Human genomics. The human transcriptome across tissues and individuals.Science. 2015 May 8;348(6235):660-5. doi: 10.1126/science.aaa0355. Science. 2015. PMID: 25954002 Free PMC article.
-
Designing a transcriptome next-generation sequencing project for a nonmodel plant species.Am J Bot. 2012 Feb;99(2):257-66. doi: 10.3732/ajb.1100292. Epub 2012 Jan 19. Am J Bot. 2012. PMID: 22268224 Review.
Cited by
-
Release of mitochondrial dsRNA into the cytosol is a key driver of the inflammatory phenotype of senescent cells.Nat Commun. 2024 Aug 27;15(1):7378. doi: 10.1038/s41467-024-51363-0. Nat Commun. 2024. PMID: 39191740 Free PMC article.
-
Comparing regional brain uptake of incretin receptor agonists after intranasal delivery in CD-1 mice and the APP/PS1 mouse model of Alzheimer's disease.Alzheimers Res Ther. 2024 Aug 1;16(1):173. doi: 10.1186/s13195-024-01537-1. Alzheimers Res Ther. 2024. PMID: 39085976 Free PMC article.
-
Inherited C-terminal TREX1 variants disrupt homology-directed repair to cause senescence and DNA damage phenotypes in Drosophila, mice, and humans.Nat Commun. 2024 Jun 1;15(1):4696. doi: 10.1038/s41467-024-49066-7. Nat Commun. 2024. PMID: 38824133 Free PMC article.
References
-
- Almanzar N, Antony J, Baghel AS, Bakerman I, Bansal I, Barres BA, Beachy PA, Berdnik D, Bilen B, Brownfield D, Cain C, Chan CKF, Chen MB, Clarke MF, Conley SD, Darmanis S, Demers A, Demir K, de Morree A, Divita T, du Bois H, Ebadi H, Espinoza FH, Fish M, Gan Q, George BM, Gillich A, Gòmez-Sjöberg R, Green F, Genetiano G, Gu X, Gulati GS, Hahn O, Haney MS, Hang Y, Harris L, He M, Hosseinzadeh S, Huang A, Huang KC, Iram T, Isobe T, Ives F, Jones RC, Kao KS, Karkanias J, Karnam G, Keller A, Kershner AM, Khoury N, Kim SK, Kiss BM, Kong W, Krasnow MA, Kumar ME, Kuo CS, Lam J, Lee DP, Lee SE, Lehallier B, Leventhal O, Li G, Li Q, Liu L, Lo A, Lu WJ, Lugo-Fagundo MF, Manjunath A, May AP, Maynard A, McGeever A, McKay M, McNerney MW, Merrill B, Metzger RJ, Mignardi M, Min D, Nabhan AN, Neff NF, Ng KM, Nguyen PK, Noh J, Nusse R, Pálovics R, Patkar R, Peng WC, Penland L, Pisco AO, Pollard K, Puccinelli R, Qi Z, Quake SR, Rando TA, Rulifson EJ, Schaum N, Segal JM, Sikandar SS, Sinha R, Sit RV, Sonnenburg J, Staehli D, Szade K, Tan M, Tan W, Tato C, Tellez K, Dulgeroff LBT, Travaglini KJ, Tropini C, Tsui M, Waldburger L, Wang BM, van Weele LJ, Weinberg K, Weissman IL, Wosczyna MN, Wu SM, Wyss-Coray T, Xiang J, Xue S, Yamauchi KA, Yang AC, Yerra LP, Youngyunpipatkul J, Yu B, Zanini F, Zardeneta ME, Zee A, Zhao C, Zhang F, Zhang H, Zhang MJ, Zhou L, Zou J, The Tabula Muris Consortium A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020;583:590–595. doi: 10.1038/s41586-020-2496-1. - DOI - PMC - PubMed
-
- Aramillo Irizar P, Schäuble S, Esser D, Groth M, Frahm C, Priebe S, Baumgart M, Hartmann N, Marthandan S, Menzel U, Müller J, Schmidt S, Ast V, Caliebe A, König R, Krawczak M, Ristow M, Schuster S, Cellerino A, Diekmann S, Englert C, Hemmerich P, Sühnel J, Guthke R, Witte OW, Platzer M, Ruppin E, Kaleta C. Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly. Nature Communications. 2018;9:327. doi: 10.1038/s41467-017-02395-2. - DOI - PMC - PubMed
-
- Benayoun BA, Pollina EA, Singh PP, Mahmoudi S, Harel I, Casey KM, Dulken BW, Kundaje A, Brunet A. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Research. 2019;29:697–709. doi: 10.1101/gr.240093.118. - DOI - PMC - PubMed
MeSH terms
Associated data
Grants and funding
- Installation Grant 3057/European Molecular Biology Organization
- FCT Investigator Starting Grant IF/00595/2014/Fundação para a Ciência e a Tecnologia
- CEEC Individual Assistant Researcher contract CEECIND/00436/2018/Fundação para a Ciência e a Tecnologia
- PhD Studentship SFRH/BD/131312/2017/Fundação para a Ciência e a Tecnologia
- PhD Studentship UI/BD/153368/2022/Fundação para a Ciência e a Tecnologia
- PhD Studentship UI/BD/153363/2022/Fundação para a Ciência e a Tecnologia
- Project LA/P/0082/2020/Fundação para a Ciência e a Tecnologia
- Project UIDP/50005/2020/Fundação para a Ciência e a Tecnologia
- Twinning grant RiboMed 857119/Horizon 2020 Framework Programme
- GenomePT project POCI-01-0145-FEDER-022184/Fundação para a Ciência e a Tecnologia
- GenomePT project POCI-01-0145-FEDER-022184/European Regional Development Fund
LinkOut - more resources
Full Text Sources

