Transcription termination and readthrough in African swine fever virus
- PMID: 38545109
- PMCID: PMC10965686
- DOI: 10.3389/fimmu.2024.1350267
Transcription termination and readthrough in African swine fever virus
Abstract
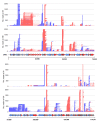
Introduction: African swine fever virus (ASFV) is a nucleocytoplasmic large DNA virus (NCLDV) that encodes its own host-like RNA polymerase (RNAP) and factors required to produce mature mRNA. The formation of accurate mRNA 3' ends by ASFV RNAP depends on transcription termination, likely enabled by a combination of sequence motifs and transcription factors, although these are poorly understood. The termination of any RNAP is rarely 100% efficient, and the transcriptional "readthrough" at terminators can generate long mRNAs which may interfere with the expression of downstream genes. ASFV transcriptome analyses reveal a landscape of heterogeneous mRNA 3' termini, likely a combination of bona fide termination sites and the result of mRNA degradation and processing. While short-read sequencing (SRS) like 3' RNA-seq indicates an accumulation of mRNA 3' ends at specific sites, it cannot inform about which promoters and transcription start sites (TSSs) directed their synthesis, i.e., information about the complete and unprocessed mRNAs at nucleotide resolution.
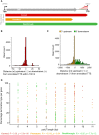
Methods: Here, we report a rigorous analysis of full-length ASFV transcripts using long-read sequencing (LRS). We systematically compared transcription termination sites predicted from SRS 3' RNA-seq with 3' ends mapped by LRS during early and late infection.
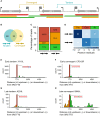
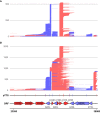
Results: Using in-vitro transcription assays, we show that recombinant ASFV RNAP terminates transcription at polyT stretches in the non-template strand, similar to the archaeal RNAP or eukaryotic RNAPIII, unaided by secondary RNA structures or predicted viral termination factors. Our results cement this T-rich motif (U-rich in the RNA) as a universal transcription termination signal in ASFV. Many genes share the usage of the same terminators, while genes can also use a range of terminators to generate transcript isoforms varying enormously in length. A key factor in the latter phenomenon is the highly abundant terminator readthrough we observed, which is more prevalent during late compared with early infection.
Discussion: This indicates that ASFV mRNAs under the control of late gene promoters utilize different termination mechanisms and factors to early promoters and/or that cellular factors influence the viral transcriptome landscape differently during the late stages of infection.
Keywords: African swine fever virus (ASFV); Oxford Nanopore; RNA polymerase; long-read sequencing; transcription readthrough; transcription termination; transcriptomics.
Copyright © 2024 Cackett, Sýkora, Portugal, Dulson, Dixon and Werner.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
The African Swine Fever Virus Transcriptome.J Virol. 2020 Apr 16;94(9):e00119-20. doi: 10.1128/JVI.00119-20. Print 2020 Apr 16. J Virol. 2020. PMID: 32075923 Free PMC article.
-
African Swine Fever Virus and Host Response: Transcriptome Profiling of the Georgia 2007/1 Strain and Porcine Macrophages.J Virol. 2022 Mar 9;96(5):e0193921. doi: 10.1128/jvi.01939-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019713 Free PMC article.
-
Transcriptome view of a killer: African swine fever virus.Biochem Soc Trans. 2020 Aug 28;48(4):1569-1581. doi: 10.1042/BST20191108. Biochem Soc Trans. 2020. PMID: 32725217 Free PMC article. Review.
-
Structure of the recombinant RNA polymerase from African Swine Fever Virus.Nat Commun. 2024 Feb 21;15(1):1606. doi: 10.1038/s41467-024-45842-7. Nat Commun. 2024. PMID: 38383525 Free PMC article.
-
African swine fever virus transcription.Virus Res. 2013 Apr;173(1):15-28. doi: 10.1016/j.virusres.2012.09.014. Epub 2012 Oct 3. Virus Res. 2013. PMID: 23041356 Review.
Cited by
-
Genetic Variations of African Swine Fever Virus: Major Challenges and Prospects.Viruses. 2024 Jun 4;16(6):913. doi: 10.3390/v16060913. Viruses. 2024. PMID: 38932205 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

