Sporadic Amyotrophic Lateral Sclerosis Skeletal Muscle Transcriptome Analysis: A Comprehensive Examination of Differentially Expressed Genes
- PMID: 38540795
- PMCID: PMC10967759
- DOI: 10.3390/biom14030377
Sporadic Amyotrophic Lateral Sclerosis Skeletal Muscle Transcriptome Analysis: A Comprehensive Examination of Differentially Expressed Genes
Abstract
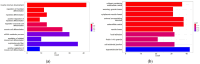
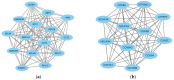
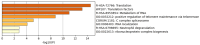
Amyotrophic lateral sclerosis (ALS) that comprises sporadic (sALS) and familial (fALS) cases, is a devastating neurodegenerative disorder characterized by progressive degeneration of motor neurons, leading to muscle atrophy and various clinical manifestations. However, the complex underlying mechanisms affecting this disease are not yet known. On the other hand, there is also no good prognosis of the disease due to the lack of biomarkers and therapeutic targets. Therefore, in this study, by means of bioinformatics analysis, sALS-affected muscle tissue was analyzed using the GEO GSE41414 dataset, identifying 397 differentially expressed genes (DEGs). Functional analysis revealed 320 up-regulated DEGs associated with muscle development and 77 down-regulated DEGs linked to energy metabolism. Protein-protein interaction network analysis identified 20 hub genes, including EIF4A1, HNRNPR and NDUFA4. Furthermore, miRNA target gene networks revealed 17 miRNAs linked to hub genes, with hsa-mir-206, hsa-mir-133b and hsa-mir-100-5p having been previously implicated in ALS. This study presents new potential biomarkers and therapeutic targets for ALS by correlating the information obtained with a comprehensive literature review, providing new potential targets to study their role in ALS.
Keywords: amyotrophic lateral sclerosis (ALS); bioinformatics; extracellular/circulating biomarkers; microRNA; neurodegenerative diseases (NDDs).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Identification of let-7f and miR-338 as plasma-based biomarkers for sporadic amyotrophic lateral sclerosis using meta-analysis and empirical validation.Sci Rep. 2022 Jan 26;12(1):1373. doi: 10.1038/s41598-022-05067-4. Sci Rep. 2022. PMID: 35082326 Free PMC article.
-
Differential Expression of MicroRNAs and Predicted Drug Target in Amyotrophic Lateral Sclerosis.J Mol Neurosci. 2023 Jun;73(6):375-390. doi: 10.1007/s12031-023-02124-z. Epub 2023 May 30. J Mol Neurosci. 2023. PMID: 37249795
-
Identification of a circulating miRNA signature in extracellular vesicles collected from amyotrophic lateral sclerosis patients.Brain Res. 2019 Apr 1;1708:100-108. doi: 10.1016/j.brainres.2018.12.016. Epub 2018 Dec 12. Brain Res. 2019. PMID: 30552897
-
New insights into the gene expression associated to amyotrophic lateral sclerosis.Life Sci. 2018 Jan 15;193:110-123. doi: 10.1016/j.lfs.2017.12.016. Epub 2017 Dec 11. Life Sci. 2018. PMID: 29241710 Review.
-
Skeletal Muscle MicroRNAs as Key Players in the Pathogenesis of Amyotrophic Lateral Sclerosis.Int J Mol Sci. 2018 May 22;19(5):1534. doi: 10.3390/ijms19051534. Int J Mol Sci. 2018. PMID: 29786645 Free PMC article. Review.
References
-
- Šoltić D., Bowerman M., Stock J., Shorrock H.K., Gillingwater T.H., Fuller H.R. Multi-Study Proteomic and Bioinformatic Identification of Molecular Overlap between Amyotrophic Lateral Sclerosis (Als) and Spinal Muscular Atrophy (Sma) Brain Sci. 2018;8:212. doi: 10.3390/brainsci8120212. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

