Exploring Transcriptional Regulation of Beta Cell SASP by Brd4-Associated Proteins and Cell Cycle Control Protein p21
- PMID: 38534794
- PMCID: PMC10968907
- DOI: 10.3390/epigenomes8010010
Exploring Transcriptional Regulation of Beta Cell SASP by Brd4-Associated Proteins and Cell Cycle Control Protein p21
Abstract
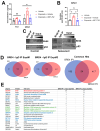
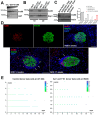
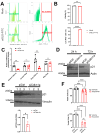
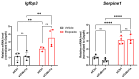
Type 1 diabetes (T1D) is a metabolic disease resulting from progressive autoimmune destruction of insulin-producing pancreatic beta cells. Although the majority of beta cells are lost in T1D, a small subset undergoes senescence, a stress response involving growth arrest, DNA damage response, and activation of a senescence-associated secretory phenotype (SASP). SASP in beta cells of the nonobese diabetic (NOD) mouse model of T1D and primary human islets is regulated at the level of transcription by bromodomain extra-terminal (BET) proteins, but the mechanisms remain unclear. To explore how SASP is transcriptionally regulated in beta cells, we used the NOD beta cell line NIT-1 to model beta cell SASP and identified binding partners of BET protein Brd4 and explored the role of the cyclin-dependent kinase inhibitor p21. Brd4 interacted with a variety of proteins in senescent NIT-1 cells including subunits of the Ino80 chromatin remodeling complex, which was expressed in beta cells during T1D progression in NOD mice and in human beta cells of control, autoantibody-positive, and T1D donors as determined from single-cell RNA-seq data. RNAi knockdown of p21 during senescence in NIT-1 cells did not significantly impact viability or SASP. Taken together, these results suggest that Brd4 interacts with several protein partners during senescence in NIT-1 cells, some of which may play roles in SASP gene activation and that p21 is dispensable for the SASP in this beta cell model.
Keywords: SASP; bromodomain extra-terminal domain proteins; cellular senescence; pancreatic beta cells; type 1 diabetes.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
BET Proteins Are Required for Transcriptional Activation of the Senescent Islet Cell Secretome in Type 1 Diabetes.Int J Mol Sci. 2019 Sep 26;20(19):4776. doi: 10.3390/ijms20194776. Int J Mol Sci. 2019. PMID: 31561444 Free PMC article.
-
DNA damage to β cells in culture recapitulates features of senescent β cells that accumulate in type 1 diabetes.Mol Metab. 2022 Aug;62:101524. doi: 10.1016/j.molmet.2022.101524. Epub 2022 Jun 2. Mol Metab. 2022. PMID: 35660116 Free PMC article.
-
Secreted GDF15 maintains transcriptional responses during DNA damage-mediated senescence in human beta cells.Am J Physiol Endocrinol Metab. 2024 Oct 1;327(4):E552-E562. doi: 10.1152/ajpendo.00257.2024. Epub 2024 Aug 28. Am J Physiol Endocrinol Metab. 2024. PMID: 39196800
-
Exploring senescence as a modifier of β cell extracellular vesicles in type 1 diabetes.Front Endocrinol (Lausanne). 2024 Aug 22;15:1422279. doi: 10.3389/fendo.2024.1422279. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39239092 Free PMC article. Review.
-
Cellular Senescence in Diabetes Mellitus: Distinct Senotherapeutic Strategies for Adipose Tissue and Pancreatic β Cells.Front Endocrinol (Lausanne). 2022 Mar 31;13:869414. doi: 10.3389/fendo.2022.869414. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35432205 Free PMC article. Review.
References
-
- Lawrence J.M., Divers J., Isom S., Saydah S., Imperatore G., Pihoker C., Marcovina S.M., Mayer-Davis E.J., Hamman R.F., Dolan L., et al. Trends in Prevalence of Type 1 and Type 2 Diabetes in Children and Adolescents in the US, 2001–2017. JAMA. 2021;326:717–727. doi: 10.1001/jama.2021.11165. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

