Porcine low-density lipoprotein receptor plays an important role in classical swine fever virus infection
- PMID: 38514916
- PMCID: PMC10962300
- DOI: 10.1080/22221751.2024.2327385
Porcine low-density lipoprotein receptor plays an important role in classical swine fever virus infection
Abstract
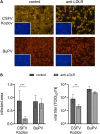
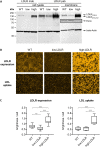
Several cellular factors have been reported to be required for replication of classical swine fever virus (CSFV), a member of the genus Pestivirus within the family Flaviviridae. However, many steps of its replication cycle are still poorly understood. The low-density lipoprotein receptor (LDLR) is involved in cell entry and post-entry processes of different viruses including other members of the Flaviviridae. In this study, the relevance of LDLR in replication of CSFV and another porcine pestivirus, Bungowannah pestivirus (BuPV), was investigated by antibody-mediated blocking of LDLR and genetically engineered porcine cell lines providing altered LDLR expression levels. An LDLR-specific antibody largely blocked infection with CSFV, but had only a minor impact on BuPV. Infections of the genetically modified cells confirmed an LDLR-dependent replication of CSFV. Compared to wild type cells, lower and higher expression of LDLR resulted in a 3.5-fold decrease or increase in viral titers already 20 h post infection. Viral titers were 25-fold increased in LDLR-overexpressing cells compared to cells with reduced LDLR expression at 72 h post infection. The varying LDLR expression levels had no clear effect on permissivity to BuPV. A decoy receptor assay using recombinant soluble LDLR provided no evidence that LDLR may function as a receptor for CSFV or BuPV. Differences in their dependency on LDLR suggest that CSFV and BuPV likely use different mechanisms to interact with their host cells. Moreover, this study reveals similarities in the replication cycles of CSFV and other members of the family Flaviviridae that are dependent on LDLR.
Keywords: Bungowannah pestivirus; CSFV; Classical swine fever virus; Flaviviridae; LDLR; low-density lipoprotein receptor.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


Similar articles
-
Porcine Complement Regulatory Protein CD46 Is a Major Receptor for Atypical Porcine Pestivirus but Not for Classical Swine Fever Virus.J Virol. 2021 Apr 12;95(9):e02186-20. doi: 10.1128/JVI.02186-20. Print 2021 Apr 12. J Virol. 2021. PMID: 33568504 Free PMC article.
-
The ESCRT-I Subunit Tsg101 Plays Novel Dual Roles in Entry and Replication of Classical Swine Fever Virus.J Virol. 2021 Feb 24;95(6):e01928-20. doi: 10.1128/JVI.01928-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33328308 Free PMC article.
-
Comparative Analysis of Tunisian Sheep-like Virus, Bungowannah Virus and Border Disease Virus Infection in the Porcine Host.Viruses. 2021 Aug 4;13(8):1539. doi: 10.3390/v13081539. Viruses. 2021. PMID: 34452404 Free PMC article.
-
Attachment, Entry, and Intracellular Trafficking of Classical Swine Fever Virus.Viruses. 2023 Sep 3;15(9):1870. doi: 10.3390/v15091870. Viruses. 2023. PMID: 37766277 Free PMC article. Review.
-
Host cell factors involved in classical swine fever virus entry.Vet Res. 2023 Dec 1;54(1):115. doi: 10.1186/s13567-023-01238-x. Vet Res. 2023. PMID: 38041163 Free PMC article. Review.
Cited by
-
Lipoprotein receptors: A little grease for enveloped viruses to open the lock?J Biol Chem. 2024 Nov;300(11):107849. doi: 10.1016/j.jbc.2024.107849. Epub 2024 Sep 30. J Biol Chem. 2024. PMID: 39357828 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
