Reciprocal inhibition between TP63 and STAT1 regulates anti-tumor immune response through interferon-γ signaling in squamous cancer
- PMID: 38509096
- PMCID: PMC10954759
- DOI: 10.1038/s41467-024-46785-9
Reciprocal inhibition between TP63 and STAT1 regulates anti-tumor immune response through interferon-γ signaling in squamous cancer
Abstract
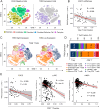
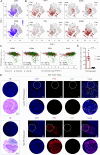
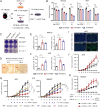
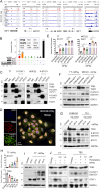
Squamous cell carcinomas (SCCs) are common and aggressive malignancies. Immune check point blockade (ICB) therapy using PD-1/PD-L1 antibodies has been approved in several types of advanced SCCs. However, low response rate and treatment resistance are common. Improving the efficacy of ICB therapy requires better understanding of the mechanism of immune evasion. Here, we identify that the SCC-master transcription factor TP63 suppresses interferon-γ (IFNγ) signaling. TP63 inhibition leads to increased CD8+ T cell infiltration and heighten tumor killing in in vivo syngeneic mouse model and ex vivo co-culture system, respectively. Moreover, expression of TP63 is negatively correlated with CD8+ T cell infiltration and activation in patients with SCC. Silencing of TP63 enhances the anti-tumor efficacy of PD-1 blockade by promoting CD8+ T cell infiltration and functionality. Mechanistically, TP63 and STAT1 mutually suppress each other to regulate the IFNγ signaling by co-occupying and co-regulating their own promoters and enhancers. Together, our findings elucidate a tumor-extrinsic function of TP63 in promoting immune evasion of SCC cells. Over-expression of TP63 may serve as a biomarker predicting the outcome of SCC patients treated with ICB therapy, and targeting TP63/STAT/IFNγ axis may enhance the efficacy of ICB therapy for this deadly cancer.
© 2024. The Author(s).
Conflict of interest statement
Y.Y.J., Y.J., J.D., S.K., D.Z. and C.Y. are named inventors on patent pending related to this study entitled “Application of Trp63 inhibition in improving the efficacy of immunotherapy for squamous cancer” (identification number: CNIPA. 202311497087.3). Other authors declare that they have no other competing interests.
Figures



Similar articles
-
Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade.J Biol Chem. 2017 Nov 10;292(45):18530-18541. doi: 10.1074/jbc.M117.808121. Epub 2017 Sep 14. J Biol Chem. 2017. PMID: 28912267 Free PMC article.
-
Squamous cell carcinomas escape immune surveillance via inducing chronic activation and exhaustion of CD8+ T Cells co-expressing PD-1 and LAG-3 inhibitory receptors.Oncotarget. 2016 Dec 6;7(49):81341-81356. doi: 10.18632/oncotarget.13228. Oncotarget. 2016. PMID: 27835902 Free PMC article.
-
Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression.Nat Commun. 2018 Sep 6;9(1):3619. doi: 10.1038/s41467-018-06081-9. Nat Commun. 2018. PMID: 30190462 Free PMC article.
-
Programmed Cell Death-Ligand-1 expression in Bladder Schistosomal Squamous Cell Carcinoma - There's room for Immune Checkpoint Blockage?.Front Immunol. 2022 Sep 2;13:955000. doi: 10.3389/fimmu.2022.955000. eCollection 2022. Front Immunol. 2022. PMID: 36148227 Free PMC article. Review.
-
IFNγ signaling integrity in colorectal cancer immunity and immunotherapy.Cell Mol Immunol. 2022 Jan;19(1):23-32. doi: 10.1038/s41423-021-00735-3. Epub 2021 Aug 12. Cell Mol Immunol. 2022. PMID: 34385592 Free PMC article. Review.
Cited by
-
Current Landscape of Cancer Immunotherapy: Harnessing the Immune Arsenal to Overcome Immune Evasion.Biology (Basel). 2024 Apr 28;13(5):307. doi: 10.3390/biology13050307. Biology (Basel). 2024. PMID: 38785789 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

