Expanding the PRAAS spectrum: De novo mutations of immunoproteasome subunit β-type 10 in six infants with SCID-Omenn syndrome
- PMID: 38503300
- PMCID: PMC11023912
- DOI: 10.1016/j.ajhg.2024.02.013
Expanding the PRAAS spectrum: De novo mutations of immunoproteasome subunit β-type 10 in six infants with SCID-Omenn syndrome
Abstract
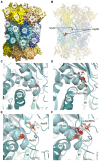
Mutations in proteasome β-subunits or their chaperone and regulatory proteins are associated with proteasome-associated autoinflammatory disorders (PRAAS). We studied six unrelated infants with three de novo heterozygous missense variants in PSMB10, encoding the proteasome β2i-subunit. Individuals presented with T-B-NK± severe combined immunodeficiency (SCID) and clinical features suggestive of Omenn syndrome, including diarrhea, alopecia, and desquamating erythematous rash. Remaining T cells had limited T cell receptor repertoires, a skewed memory phenotype, and an elevated CD4/CD8 ratio. Bone marrow examination indicated severely impaired B cell maturation with limited V(D)J recombination. All infants received an allogeneic stem cell transplant and exhibited a variety of severe inflammatory complications thereafter, with 2 peri-transplant and 2 delayed deaths. The single long-term transplant survivor showed evidence for genetic rescue through revertant mosaicism overlapping the affected PSMB10 locus. The identified variants (c.166G>C [p.Asp56His] and c.601G>A/c.601G>C [p.Gly201Arg]) were predicted in silico to profoundly disrupt 20S immunoproteasome structure through impaired β-ring/β-ring interaction. Our identification of PSMB10 mutations as a cause of SCID-Omenn syndrome reinforces the connection between PRAAS-related diseases and SCID.
Keywords: Omenn syndrome; PSMB10; immunoproteasome; revertant somatic mosaicism; severe combined immune deficiency; uniparental disomy.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Similar articles
-
Heterozygous missense variant of the proteasome subunit β-type 9 causes neonatal-onset autoinflammation and immunodeficiency.Nat Commun. 2021 Nov 24;12(1):6819. doi: 10.1038/s41467-021-27085-y. Nat Commun. 2021. PMID: 34819510 Free PMC article.
-
Molecular analysis of T-B-NK+ severe combined immunodeficiency and Omenn syndrome cases in Saudi Arabia.BMC Med Genet. 2009 Nov 13;10:116. doi: 10.1186/1471-2350-10-116. BMC Med Genet. 2009. PMID: 19912631 Free PMC article.
-
Identification of eight novel proteasome variants in five unrelated cases of proteasome-associated autoinflammatory syndromes (PRAAS).Front Immunol. 2023 Aug 4;14:1190104. doi: 10.3389/fimmu.2023.1190104. eCollection 2023. Front Immunol. 2023. PMID: 37600812 Free PMC article.
-
Omenn syndrome does not live by V(D)J recombination alone.Curr Opin Allergy Clin Immunol. 2011 Dec;11(6):525-31. doi: 10.1097/ACI.0b013e32834c311a. Curr Opin Allergy Clin Immunol. 2011. PMID: 22001740 Review.
-
Omenn syndrome--review of several phenotypes of Omenn syndrome and RAG1/RAG2 mutations in Japan.Allergol Int. 2006 Jun;55(2):115-9. doi: 10.2332/allergolint.55.115. Allergol Int. 2006. PMID: 17075247 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

