Exploring associations between the teat apex metagenome and Staphylococcus aureus intramammary infections in primiparous cows under organic directives
- PMID: 38497641
- PMCID: PMC11022539
- DOI: 10.1128/aem.02234-23
Exploring associations between the teat apex metagenome and Staphylococcus aureus intramammary infections in primiparous cows under organic directives
Abstract
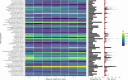
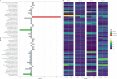
The primary objective of this study was to identify associations between the prepartum teat apex microbiome and the presence of Staphylococcus aureus intramammary infections (IMI) in primiparous cows during the first 5 weeks after calving. We performed a case-control study using shotgun metagenomics of the teat apex and culture-based milk data collected longitudinally from 710 primiparous cows on five organic dairy farms. Cases had higher odds of having S. aureus metagenomic DNA on the teat apex prior to parturition compared to controls (OR = 38.9, 95% CI: 14.84-102.21). Differential abundance analysis confirmed this association, with cases having a 23.8 higher log fold change (LFC) in the abundance of S. aureus in their samples compared to controls. Of the most prevalent microorganisms in controls, those associated with a lower risk of post-calving S. aureus IMI included Microbacterium phage Min 1 (OR = 0.37, 95% CI: 0.25-0.53), Corynebacterium efficiens (OR = 0.53, 95% CI: 0.30-0.94), Kocuria polaris (OR = 0.54, 95% CI: 0.35-0.82), Micrococcus terreus (OR = 0.64, 95% CI: 0.44-0.93), and Dietzia alimentaria (OR = 0.45, 95% CI: 0.26-0.75). Genes encoding for Microcin B17 AMPs were the most prevalent on the teat apex of cases and controls (99.7% in both groups). The predicted abundance of genes encoding for Microcin B17 was also higher in cases compared to controls (LFC 0.26).
Importance: Intramammary infections (IMI) caused by Staphylococcus aureus remain an important problem for the dairy industry. The microbiome on the external skin of the teat apex may play a role in mitigating S. aureus IMI risk, in particular the production of antimicrobial peptides (AMPs) by commensal microbes. However, current studies of the teat apex microbiome utilize a 16S approach, which precludes the detection of genomic features such as genes that encode for AMPs. Therefore, further research using a shotgun metagenomic approach is needed to understand what role prepartum teat apex microbiome dynamics play in IMI risk.
Keywords: metagenomics; microbial ecology; veterinary epidemiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Investigation of intramammary infections in primiparous cows during early lactation on organic dairy farms.J Dairy Sci. 2023 Dec;106(12):9377-9392. doi: 10.3168/jds.2022-23036. Epub 2023 Aug 23. J Dairy Sci. 2023. PMID: 37641314
-
The influence of the rearing period on intramammary infections in Swiss dairy heifers: A cross-sectional study.Prev Vet Med. 2016 Jul 1;129:23-34. doi: 10.1016/j.prevetmed.2016.04.013. Epub 2016 May 13. Prev Vet Med. 2016. PMID: 27317320
-
Communications of Staphylococcus aureus and non-aureus Staphylococcus species from bovine intramammary infections and teat apex colonization.J Dairy Sci. 2018 Aug;101(8):7322-7333. doi: 10.3168/jds.2017-14311. Epub 2018 May 16. J Dairy Sci. 2018. PMID: 29778469
-
Association between teat skin colonization and intramammary infection with Staphylococcus aureus and Streptococcus agalactiae in herds with automatic milking systems.J Dairy Sci. 2019 Jan;102(1):629-639. doi: 10.3168/jds.2018-15330. Epub 2018 Nov 8. J Dairy Sci. 2019. PMID: 30415854
-
Associations between early lactation intramammary infections and udder health and performance during the first 180 days in milk in first-lactation organic dairy cows.J Dairy Sci. 2024 Apr;107(4):2426-2443. doi: 10.3168/jds.2023-23924. Epub 2023 Nov 2. J Dairy Sci. 2024. PMID: 37923212
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

