This is a preprint.
A tale of two fusion proteins: understanding the metastability of human respiratory syncytial virus and metapneumovirus and implications for rational design of uncleaved prefusion-closed trimers
- PMID: 38496645
- PMCID: PMC10942449
- DOI: 10.1101/2024.03.07.583986
A tale of two fusion proteins: understanding the metastability of human respiratory syncytial virus and metapneumovirus and implications for rational design of uncleaved prefusion-closed trimers
Update in
-
Rational design of uncleaved prefusion-closed trimer vaccines for human respiratory syncytial virus and metapneumovirus.Nat Commun. 2024 Nov 16;15(1):9939. doi: 10.1038/s41467-024-54287-x. Nat Commun. 2024. PMID: 39550381 Free PMC article.
Abstract
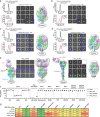
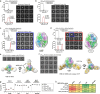
Respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) cause human respiratory diseases and are major targets for vaccine development. In this study, we designed uncleaved prefusion-closed (UFC) trimers for the fusion (F) proteins of both viruses by examining mutations critical to F metastability. For RSV, we assessed four previous prefusion F designs, including the first and second generations of DS-Cav1, SC-TM, and 847A. We then identified key mutations that can maintain prefusion F in a native-like, closed trimeric form (up to 76%) without introducing any interprotomer disulfide bond. For hMPV, we developed a stable UFC trimer with a truncated F2-F1 linkage and an interprotomer disulfide bond. Tens of UFC constructs were characterized by negative-stain electron microscopy (nsEM), x-ray crystallography (11 RSV-F and one hMPV-F structures), and antigenic profiling. Using an optimized RSV-F UFC trimer as bait, we identified three potent RSV neutralizing antibodies (NAbs) from a phage-displayed human antibody library, with a public NAb lineage targeting sites Ø and V and two cross-pneumovirus NAbs recognizing site III. In mouse immunization, rationally designed RSV-F and hMPV-F UFC trimers induced robust antibody responses with high neutralizing titers. Our study provides a foundation for future prefusion F-based RSV and hMPV vaccine development.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures







Similar articles
-
Rational design of uncleaved prefusion-closed trimer vaccines for human respiratory syncytial virus and metapneumovirus.Nat Commun. 2024 Nov 16;15(1):9939. doi: 10.1038/s41467-024-54287-x. Nat Commun. 2024. PMID: 39550381 Free PMC article.
-
A monomeric uncleaved respiratory syncytial virus F antigen retains prefusion-specific neutralizing epitopes.J Virol. 2014 Oct;88(20):11802-10. doi: 10.1128/JVI.01225-14. Epub 2014 Jul 30. J Virol. 2014. PMID: 25078705 Free PMC article.
-
Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses.Proc Natl Acad Sci U S A. 2021 Sep 28;118(39):e2106196118. doi: 10.1073/pnas.2106196118. Proc Natl Acad Sci U S A. 2021. PMID: 34551978 Free PMC article.
-
Antibody Epitopes of Pneumovirus Fusion Proteins.Front Immunol. 2019 Nov 29;10:2778. doi: 10.3389/fimmu.2019.02778. eCollection 2019. Front Immunol. 2019. PMID: 31849961 Free PMC article. Review.
-
Structural basis for respiratory syncytial virus and human metapneumovirus neutralization.Curr Opin Virol. 2023 Aug;61:101337. doi: 10.1016/j.coviro.2023.101337. Curr Opin Virol. 2023. PMID: 37544710 Free PMC article. Review.
References
-
- Falsey A. R., Hennessey P. A., Formica M. A., Cox C., Walsh E. E., Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352, 1749–1759 (2005). - PubMed
-
- Stockman L. J., Curns A. T., Anderson L. J., Fischer-Langley G., Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997–2006. Pediatr. Infect. Dis. J. 31, 5–9 (2012). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
