VirtuousPocketome: a computational tool for screening protein-ligand complexes to identify similar binding sites
- PMID: 38491261
- PMCID: PMC10943019
- DOI: 10.1038/s41598-024-56893-7
VirtuousPocketome: a computational tool for screening protein-ligand complexes to identify similar binding sites
Abstract
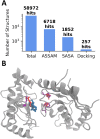
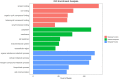
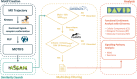
Protein residues within binding pockets play a critical role in determining the range of ligands that can interact with a protein, influencing its structure and function. Identifying structural similarities in proteins offers valuable insights into their function and activation mechanisms, aiding in predicting protein-ligand interactions, anticipating off-target effects, and facilitating the development of therapeutic agents. Numerous computational methods assessing global or local similarity in protein cavities have emerged, but their utilization is impeded by complexity, impractical automation for amino acid pattern searches, and an inability to evaluate the dynamics of scrutinized protein-ligand systems. Here, we present a general, automatic and unbiased computational pipeline, named VirtuousPocketome, aimed at screening huge databases of proteins for similar binding pockets starting from an interested protein-ligand complex. We demonstrate the pipeline's potential by exploring a recently-solved human bitter taste receptor, i.e. the TAS2R46, complexed with strychnine. We pinpointed 145 proteins sharing similar binding sites compared to the analysed bitter taste receptor and the enrichment analysis highlighted the related biological processes, molecular functions and cellular components. This work represents the foundation for future studies aimed at understanding the effective role of tastants outside the gustatory system: this could pave the way towards the rationalization of the diet as a supplement to standard pharmacological treatments and the design of novel tastants-inspired compounds to target other proteins involved in specific diseases or disorders. The proposed pipeline is publicly accessible, can be applied to any protein-ligand complex, and could be expanded to screen any database of protein structures.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The human bitter taste receptor TAS2R10 is tailored to accommodate numerous diverse ligands.J Neurosci. 2013 Jan 2;33(1):201-13. doi: 10.1523/JNEUROSCI.3248-12.2013. J Neurosci. 2013. PMID: 23283334 Free PMC article.
-
Dual binding mode of "bitter sugars" to their human bitter taste receptor target.Sci Rep. 2019 Jun 11;9(1):8437. doi: 10.1038/s41598-019-44805-z. Sci Rep. 2019. PMID: 31186454 Free PMC article.
-
Ligand binding modes from low resolution GPCR models and mutagenesis: chicken bitter taste receptor as a test-case.Sci Rep. 2017 Aug 15;7(1):8223. doi: 10.1038/s41598-017-08344-9. Sci Rep. 2017. PMID: 28811548 Free PMC article.
-
New insights into the characteristics of sweet and bitter taste receptors.Int Rev Cell Mol Biol. 2011;291:191-226. doi: 10.1016/B978-0-12-386035-4.00006-9. Int Rev Cell Mol Biol. 2011. PMID: 22017977 Review.
-
The heterodimeric sweet taste receptor has multiple potential ligand binding sites.Curr Pharm Des. 2006;12(35):4591-600. doi: 10.2174/138161206779010350. Curr Pharm Des. 2006. PMID: 17168764 Review.
Cited by
-
Exploring Cannabinoids as Potential Inhibitors of SARS-CoV-2 Papain-like Protease: Insights from Computational Analysis and Molecular Dynamics Simulations.Viruses. 2024 May 30;16(6):878. doi: 10.3390/v16060878. Viruses. 2024. PMID: 38932170 Free PMC article.
-
Single-Nucleotide Polymorphisms of TAS2R46 Affect the Receptor Downstream Calcium Regulation in Histamine-Challenged Cells.Cells. 2024 Jul 16;13(14):1204. doi: 10.3390/cells13141204. Cells. 2024. PMID: 39056786 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

