Revealing the molecular landscape of human placenta: a systematic review and meta-analysis of single-cell RNA sequencing studies
- PMID: 38478759
- PMCID: PMC11215163
- DOI: 10.1093/humupd/dmae006
Revealing the molecular landscape of human placenta: a systematic review and meta-analysis of single-cell RNA sequencing studies
Abstract
Background: With increasing significance of developmental programming effects associated with placental dysfunction, more investigations are devoted to improving the characterization and understanding of placental signatures in health and disease. The placenta is a transitory but dynamic organ adapting to the shifting demands of fetal development and available resources of the maternal supply throughout pregnancy. Trophoblasts (cytotrophoblasts, syncytiotrophoblasts, and extravillous trophoblasts) are placental-specific cell types responsible for the main placental exchanges and adaptations. Transcriptomic studies with single-cell resolution have led to advances in understanding the placenta's role in health and disease. These studies, however, often show discrepancies in characterization of the different placental cell types.
Objective and rationale: We aim to review the knowledge regarding placental structure and function gained from the use of single-cell RNA sequencing (scRNAseq), followed by comparing cell-type-specific genes, highlighting their similarities and differences. Moreover, we intend to identify consensus marker genes for the various trophoblast cell types across studies. Finally, we will discuss the contributions and potential applications of scRNAseq in studying pregnancy-related diseases.
Search methods: We conducted a comprehensive systematic literature review to identify different cell types and their functions at the human maternal-fetal interface, focusing on all original scRNAseq studies on placentas published before March 2023 and published reviews (total of 28 studies identified) using PubMed search. Our approach involved curating cell types and subtypes that had previously been defined using scRNAseq and comparing the genes used as markers or identified as potential new markers. Next, we reanalyzed expression matrices from the six available scRNAseq raw datasets with cell annotations (four from first trimester and two at term), using Wilcoxon rank-sum tests to compare gene expression among studies and annotate trophoblast cell markers in both first trimester and term placentas. Furthermore, we integrated scRNAseq raw data available from 18 healthy first trimester and nine term placentas, and performed clustering and differential gene expression analysis. We further compared markers obtained with the analysis of annotated and raw datasets with the literature to obtain a common signature gene list for major placental cell types.
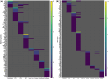
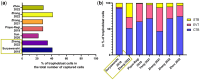
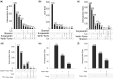
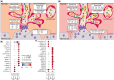
Outcomes: Variations in the sampling site, gestational age, fetal sex, and subsequent sequencing and analysis methods were observed between the studies. Although their proportions varied, the three trophoblast types were consistently identified across all scRNAseq studies, unlike other non-trophoblast cell types. Notably, no marker genes were shared by all studies for any of the investigated cell types. Moreover, most of the newly defined markers in one study were not observed in other studies. These discrepancies were confirmed by our analysis on trophoblast cell types, where hundreds of potential marker genes were identified in each study but with little overlap across studies. From 35 461 and 23 378 cells of high quality in the first trimester and term placentas, respectively, we obtained major placental cell types, including perivascular cells that previously had not been identified in the first trimester. Importantly, our meta-analysis provides marker genes for major placental cell types based on our extensive curation.
Wider implications: This review and meta-analysis emphasizes the need for establishing a consensus for annotating placental cell types from scRNAseq data. The marker genes identified here can be deployed for defining human placental cell types, thereby facilitating and improving the reproducibility of trophoblast cell annotation.
Keywords: cell type annotation; marker genes; single-cell RNA sequencing; single-nucleus RNA sequencing; trophoblasts.
© The Author(s) 2024. Published by Oxford University Press on behalf of European Society of Human Reproduction and Embryology.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

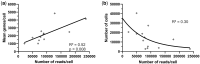


Similar articles
-
TGFβ signalling: a nexus between inflammation, placental health and preeclampsia throughout pregnancy.Hum Reprod Update. 2024 Jul 1;30(4):442-471. doi: 10.1093/humupd/dmae007. Hum Reprod Update. 2024. PMID: 38519450 Free PMC article. Review.
-
Current approaches and developments in transcript profiling of the human placenta.Hum Reprod Update. 2020 Nov 1;26(6):799-840. doi: 10.1093/humupd/dmaa028. Hum Reprod Update. 2020. PMID: 33043357 Free PMC article. Review.
-
Human Placental Endothelial Cell and Trophoblast Heterogeneity and Differentiation Revealed by Single-Cell RNA Sequencing.Cells. 2022 Dec 25;12(1):87. doi: 10.3390/cells12010087. Cells. 2022. PMID: 36611882 Free PMC article.
-
Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta.Cell Res. 2018 Aug;28(8):819-832. doi: 10.1038/s41422-018-0066-y. Epub 2018 Jul 24. Cell Res. 2018. PMID: 30042384 Free PMC article.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
Cited by
-
Placental Origins of Preeclampsia: Insights from Multi-Omic Studies.Int J Mol Sci. 2024 Aug 28;25(17):9343. doi: 10.3390/ijms25179343. Int J Mol Sci. 2024. PMID: 39273292 Free PMC article. Review.
-
Single-cell and spatial transcriptomics reveal alterations in trophoblasts at invasion sites and disturbed myometrial immune microenvironment in placenta accreta spectrum disorders.Biomark Res. 2024 Jun 3;12(1):55. doi: 10.1186/s40364-024-00598-6. Biomark Res. 2024. PMID: 38831319 Free PMC article.
References
-
- Adu-Gyamfi EA, Lamptey J, Chen X-M, Li F-F, Li C, Ruan L-L, Yang X-N, Liu T-H, Wang Y-X, Ding Y-B. et al. Iodothyronine deiodinase 2 (DiO2) regulates trophoblast cell line cycle, invasion and apoptosis; and its downregulation is associated with early recurrent miscarriage. Placenta 2021;111:54–68. - PubMed
-
- Al-Lamki RS, Skepper JN, Burton GJ.. Are human placental bed giant cells merely aggregates of small mononuclear trophoblast cells? An ultrastructural and immunocytochemical study. Hum Reprod 1999;14:496–504. - PubMed
-
- Anacker J, Segerer SE, Hagemann C, Feix S, Kapp M, Bausch R, Kämmerer U.. Human decidua and invasive trophoblasts are rich sources of nearly all human matrix metalloproteinases. Mol Hum Reprod 2011;17:637–652. - PubMed
-
- Aplin JD. Developmental cell biology of human villous trophoblast: current research problems. Int J Dev Biol 2010;54:323–329. - PubMed
-
- Aplin JD, Haigh T, Vicovac L, Church HJ, Jones CJP.. Anchorage in the developing placenta: an overlooked determinant of pregnancy outcome? Hum Fertil (Camb) 1998;1:75–79. - PubMed

