The Impact of Mutational Hotspots on Cancer Survival
- PMID: 38473427
- PMCID: PMC10931094
- DOI: 10.3390/cancers16051072
The Impact of Mutational Hotspots on Cancer Survival
Abstract
Background: Cofactors, biomarkers, and the mutational status of genes such as TP53, EGFR, IDH1/2, or PIK3CA have been used for patient stratification. However, many genes exhibit recurrent mutational positions known as hotspots, specifically linked to varying degrees of survival outcomes. Nevertheless, few hotspots have been analyzed (e.g., TP53 and EGFR). Thus, many other genes and hotspots remain unexplored.
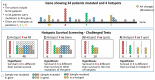
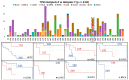
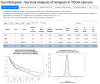
Methods: We systematically screened over 1400 hotspots across 33 TCGA cancer types. We compared the patients carrying a hotspot against (i) all cases, (ii) gene-mutated cases, (iii) other mutated hotspots, or (iv) specific hotspots. Due to the limited number of samples in hotspots and the inherent group imbalance, besides Cox models and the log-rank test, we employed VALORATE to estimate their association with survival precisely.
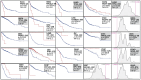
Results: We screened 1469 hotspots in 6451 comparisons, where 314 were associated with survival. Many are discussed and linked to the current literature. Our findings demonstrate associations between known hotspots and survival while also revealing more potential hotspots. To enhance accessibility and promote further investigation, all the Kaplan-Meier curves, the log-rank tests, Cox statistics, and VALORATE-estimated null distributions are accessible on our website.
Conclusions: Our analysis revealed both known and putatively novel hotspots associated with survival, which can be used as biomarkers. Our web resource is a valuable tool for cancer research.
Keywords: TCGA; VALORATE; biomarkers; cox; log-rank; recurrent mutations.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Clinical significance of frequent somatic mutations detected by high-throughput targeted sequencing in archived colorectal cancer samples.J Transl Med. 2016 May 4;14(1):118. doi: 10.1186/s12967-016-0878-9. J Transl Med. 2016. PMID: 27146902 Free PMC article.
-
VALORATE: fast and accurate log-rank test in balanced and unbalanced comparisons of survival curves and cancer genomics.Bioinformatics. 2017 Jun 15;33(12):1900-1901. doi: 10.1093/bioinformatics/btx080. Bioinformatics. 2017. PMID: 28186225
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
-
Computational methods for detecting cancer hotspots.Comput Struct Biotechnol J. 2020 Nov 19;18:3567-3576. doi: 10.1016/j.csbj.2020.11.020. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33304455 Free PMC article. Review.
-
Theoretical analysis of mutation hotspots and their DNA sequence context specificity.Mutat Res. 2003 Sep;544(1):65-85. doi: 10.1016/s1383-5742(03)00032-2. Mutat Res. 2003. PMID: 12888108 Review.
References
-
- Sugio K., Uramoto H., Ono K., Oyama T., Hanagiri T., Sugaya M., Ichiki Y., So T., Nakata S., Morita M., et al. Mutations within the Tyrosine Kinase Domain of EGFR Gene Specifically Occur in Lung Adenocarcinoma Patients with a Low Exposure of Tobacco Smoking. Br. J. Cancer. 2006;94:896–903. doi: 10.1038/sj.bjc.6603040. - DOI - PMC - PubMed
-
- Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.A., Mooij T.M., Roos-Blom M.J., Jervis S., Van Leeuwen F.E., Milne R.L., Andrieu N., et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA-J. Am. Med. Assoc. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

